High-Resolution Physical and Transcriptional Mapping of the Autoimmune Polyendocrinopathy–Candidiasis–Ectodermal Dystrophy Locus on Chromosome 21q22.3 by FISH
- Johanna Aaltonen1,5,
- Nina Horelli-Kuitunen2,5,
- Jian-Bing Fan3,
- Petra Björses1,
- Jaakko Perheentupa4,
- Richard Myers3,
- Aarno Palotie2, and
- Leena Peltonen1,6
- 1Department of Human Molecular Genetics, National Public Health Institute, 00300 Helsinki, Finland; 2Departments of Clinical Chemistry and Biomedicine, University of Helsinki, 00280 Helsinki, Finland; 3Department of Genetics and the Stanford Human Genome Center, Stanford University School of Medicine, Stanford, California 94305; 4Hospital for Children and Adolescents, University of Helsinki, 00280 Helsinki, Finland
Abstract
Autoimmune–polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED, PGD type I) is an autosomal recessive disease enriched in the Finnish population. Previously, we have assigned APECED to a 2.6-cM interval on chromosome 21q22.3 by linkage analysis in 14 Finnish families. This subtelomeric region of 21q22.3 seems to have sequence features resulting in its under-representation in large insert genomic libraries, and only a few large insert clones have been available for positional cloning to date. Here, we report the refined localization of the APECED gene and a visual physical map of 800 kb covering the critical chromosomal region for the gene. In the construction of the physical map, the recently developed fiber FISH techniques were essential for the orientation of the cosmid P1, PAC, and BAC clones in relation to each other. We also localized two cDNAs within this genomic region by fiber FISH combined with the highly sensitive tyramide-based detection method. These data will facilitate the final cloning of theAPECED gene and any other novel gene in this complex genomic region.
[On-line supplement for primer sequences, PCR product size, and annealing temperatures is available athttp://www.cshl.org/gr.]
Chromosome 21 has been the model chromosome for the Human Genome Project because of its small size. Furthermore, intensive research focused on the Down syndrome, as well as chromosome 21-linked genetic diseases, has provided detailed information on this chromosome, leading to the publication of the first physical map in 1992 (Chumakov et al. 1992). However, some gaps still exist in the physical map, particularly around the region of 21q22.3, which is suspected to represent an exceptionally gene-rich region of the human genome (Gardiner 1996).
Autoimmune–polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED, PGD type I; MIM no. 240300) is an autosomal recessive systemic autoimmune disease enriched in the Finnish population with an incidence of 1:25,000 (Ahonen et al. 1990). Another population with a relatively high frequency of the disease is the Iranian Jewish population (Zlotogora and Shapiro 1992). The disease usually begins in childhood, but new tissue-specific symptoms may appear throughout life. The classical triad of symptoms include hypoparathyroidism, primary adrenocortical failure, and chronic mucocutaneous candidiasis. Other endocrine deficiences such as diabetes mellitus, hypothyroidism, gonadal failure, and gastric parietal cell failure can occur in variable order and with variable severity among APECED patients. Evidence of involvement of tissues of ectodermal origin is characterized by signs including dystrophy of dental enamel and nails as well as alopecia and vitiligo. Previously, we have mapped the APECED locus to chromosome 21q22.3 by linkage studies in 14 Finnish families (Aaltonen et al. 1994). We have further demonstrated that non-Finnish families tested so far, including the families from the Iranian Jewish population, are linked with the same locus (Björses et al. 1996). The initial linkage data assigned the APECED locus between the polymorphic DNA markersD21S49 and D21S171 which are genetically 2.6 cM apart.
Here, we have restricted the disease locus by recombination and linkage disequilibrium analyses between markers D21S25 andD21S171 within a 350-kb chromosomal region determined by fiber fluorescence in situ hybridization (FISH). We also report a bacterial clone contig covering the DNA region critical for the cloning of the APECED gene. To achieve this, we utilized different applications of the FISH technology: a high resolution fiber FISH method in building the clone contig over this genomic region avoiding the need for more tedious pulse-field gel electrophoresis (PFGE)-based mapping (Florijn et al. 1995; Heiskanen et al. 1995); and a tyramide-based detection method, highly sensitive in assigning and visualizing small candidate cDNAs (Raap et al. 1995). Most importantly, we have provided material for future efforts in unraveling the novel genes in this gene-rich region by cDNA selection or exon trapping techniques (Gardiner et al. 1995; Yaspo et al. 1995). Also, large-scale sequencing efforts aimed at sequencing the complete chromosome 21 will benefit from our data.
RESULTS
Restricting the Critical Chromosomal Region for APECED
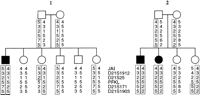
No recombinations were observed in APECED families with a highly informative dinucleotide repeat polymorphism, D21S1912, or a biallelic restriction fragment-length polymorphism (RFLP) markerD21S25. In one Finnish family, however, an ancient recombination in the haplotype was observed with the markerD21S25, further restricting the critical DNA region to between markers D21S25 and D21S171 (Fig.1). No recombinations were observed with the poorly informative intragenic markers for the PFKL locus or with the RFLP marker D21S154.
Extended haplotypes around the APECED region. The disease chromosomes are enclosed in boxes. In family 1, an ancient recombination in the haplotype was observed between D21S25and PFKL, and in family 2, an obligatory recombination was observed with the marker D21S171. Thus, the APECED locus was restricted between markersD21S25 and D21S171.
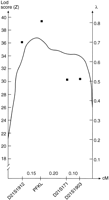
The linkage disequilibrium analyses supported the results of the haplotype analyses, suggesting restriction of the APECEDregion to between markers D21S25 and D21S171(Fig. 2). The P excess values, expressing the linkage disequilibrium, were 0.78, 0.85, and 0.60 for markers D21S1912, PFKL, and D21S171 in the Finnish family material, respectively. Only multiallelic markers were used in the linkage disequilibrium analysis because of the very low informativeness of the RFLP markers (D21S25,PFKL, and D21S154) in our family material. To further utilize linkage disequilibrium to restrict the DNA region for the APECED gene, new polymorphic repeats were searched for on the physical clones over the region using di-, tri-, and tetranucleotide repeats as probes. Surprisingly, no new highly polymorphic repeats could be identified in the region flanked by markers D21S25 and D21S171.
Multipoint association statistic for the position of theAPECED gene. The graph shows the most likely location of theAPECED gene relative to the known fixed positions of the marker loci. The estimated values of λ (proportion of excess of a certain allele of a marker in chromosomes carrying the disease allele) for each marker are indicated (▪) (Terwilliger et al. 1995).
Screening the Genomic Libraries
Table 1 summarizes the STS content for each clone. The large insert genomic libraries [chromosome 21-specific cosmid library, total genomic P1, P1-derived artificial chromosome (PAC), and bacterial artificial chromosome (BAC) libraries] were first screened by PCR with previously published markers D21S1912, PFKL, and D21S171, a sequence-tagged site (STS) for D21S25, and linking cloneD21S400 (Ichikawa et al. 1993). Markers D21S1912and D21S25 were present in the PAC 225O17 and BAC clones 175D10, 134A17, and 282P11. Clones P1 1048H3, PAC 92C23, and BAC 127P21 were positive with PFKL and D21S400. In addition, P1 1048H3, PAC 92C23, and BAC 54L7 contained D21S154, whereas BAC 127P21 did not. Another clone, P1 579E2, was positive with D21S400 andD21S154; the BAC 54L7 contained these markers as well. The marker D21S171 was present in PAC clone 314N7 and BAC clones 165H12, 122D2, 380J5, and 4D22. Two additional P1 clones, 1084E5 and 1406C8, contained D21S171.
STS Content of 21q22.3 Physical Clones Over the APECEDRegion
In addition to known STSs, two novel STSs (18744 and 23181) sequenced from both end fragments of PAC 314N7 were used to screen the genomic libraries. End fragments of the clones PAC 92C23 and P1 579E2 were also isolated, but all of them were Alu or LINE repetitive sequences and could not be utilized in the characterization of new clones. Two STSs (23937 and 23938) were sequenced from P1 124G1. The end fragment of BAC 86O23 was also used as an STS (86O23.sp6). The BAC clones 54L7 and 4D22 and P1 clone 1226M10 contained the STS 86O23.sp6. The STSs 23938 and 23937 were both present in BAC clones 4D22, 86O23, and 380J5 and P1 clones 990D10, 3E11, and 1302A3, whereas BAC clone 54L7 and P1 clone 1226M10 contained only STS 23938; STS 23937 was present only in P1 clones 1084E5 and 1406C8. The STS 18744 was present in several P1 clones: 1084E5, 1406C8, 1302A3, and 124G1. BAC clones 4D22, 380J5, 165H12, and 122D2 were also positive with this STS. PAC clone 225L15 contained the STS 23181 and the markerD21S1903. BAC clones 5O2 and 7B7 contained the STS Z500C, and BAC clone 7B7 contained CD18 as well.
Visual Mapping by FISH
Conventional FISH analysis on metaphase chromosomes, providing a mapping resolution of 2–3 Mb, was used to assign the clones to a specific chromosomal band. FISH on metaphase chromosomes was also used to exclude potential chimerism of the clones. Altogether, 52 different-sized clones were hybridized, and 30 metaphases of each preparation were analyzed. All of the clones used in this study mapped to chromosome 21q22.3 alone. The resolution of FISH analysis on metaphase chromosomes was inadequate for orientating the clones with respect to each other.
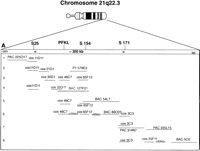
Five cosmid clones (11D11, 46C7, 65F12, 3C3, 25F11) served as anchoring markers in the fiber FISH map construction. Cosmid clone 11D11 contained the markers D21S1912 and D21S25; cosmid 46C7 contained PFKL; cosmid 65F12 was positive withD21S154; cosmid clone 3C3 contained two STSs, 23937, and 18744, and marker D21S171; and cosmid 25F11 contained STS 23181 and marker D21S1903. Each hybridization was performed with at least three probes to enable reliable determination of the order and orientation of the clones. To assure the location and distance from other probes, each probe was hybridized in combination with two to four different probes. Ten to 25 images from each hybridization were analyzed. The main anchoring probe, the cosmid clone 65F12 (39 kb), was used as a standard for distance measurements. The minimal tiling pathway, determined by fiber FISH, was as follows: centromere–PAC 225O17–cos 11D11–cos 21D1–cos 30D1–cos 22G11–BAC 127P21–P1 579E2–BAC 54L7–BAC 86O23–P1 124G1–PAC 314N7–PAC 225L15–BAC 5O2–BAC 7B7–telomere (Fig. 3A,B). This order agrees completely with the order obtained by STS content mapping (Table 1). The overlap cos 21D1–cos 30D1–cos 22G11 was also confirmed with restriction enzyme digests and subsequent agarose gel runs because of the lack of STSs in this region.
The physical map of ∼800 kb on chromosome 21q22.3 including the flanking markers D21S25 and D21S171 for theAPECED critical region. The clones shown here represent the minimal tiling pathway. (A) A graphical illustration of the clone contig and interclone distances determined by fiber FISH. (B) The fiber FISH images, with each line (1–8) representing the corresponding line in A and demonstrating one FISH image.
The last three lines of Table 1 show the number of different clone types identified per each marker and STS, reflecting the redundancy of a particular genomic library. The mean insert size was 70 kb in the P1 clones, 120 kb in the PAC clones, 110 kb in the BAC clones from Research Genetics, Inc., and 200 kb in the BAC clones from Genome Systems, Inc. Results of the distance measurements were uniform, when the distance ranged from 1 to 200 kb, but the variation between individual measurements became larger when the distance was >200 kb.
Novel Expressed Sequence Tags in the Region
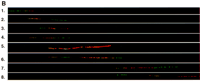
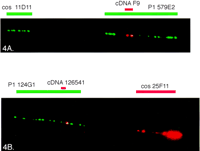
Cloned cDNAs from a cDNA selection experiment were hybridized with PAC 92C23 and also with the cosmid contig overlapping with PAC 92C23 (Stone et al. 1996). The selected cDNA F9 (size 1.0 kb) hybridized with PAC 92C23 and the cosmids 46C7, 1B4, and 5E4. We were also able to show that clone P1 579E2 contained the cDNA F9 by applying a highly sensitive tyramide-based detection method in the FISH analysis (Raap et al. 1995). The cosmid 11D11 was hybridized simultaneously with the cDNA and P1 579E2 to reveal the right orientation of the clones (Fig.4A). With the tyramine-based detection method we localized another cDNA, IMAGE clone 126541, to the telomeric side of clone P1 124G1, and cosmid 25F11 served as a marker for centromere–telomere orientation (Fig. 4B). Database searches with the sequences obtained from these cDNAs revealed no homology with known genes. However, two homologous expressed sequence tags (ESTs) were identified: The cDNA F9 was homologous with EST 21ES212, and the IMAGE clone 126541 was homologous with EST 430058.
The fiber FISH image of the region between markers D21S25and D21S154 (A) and around markerD21S171 (B). (A) A biotin-labeled candidate cDNA F9 (red) is visualized by tyramide detection on P1 clone 579E2 (long green signal), and a digoxigenin-labeled cosmid (cos) 11D11 (green) shows the orientation of the clones (centromere–cos 11D11–P1 579E2/cDNA F9–telomere). (B) A biotin-labeled IMAGE clone 126541 (red) is positioned on P1 124G1 (green) and cosmid 25F11 is visualized by tyramide-based detection to confirm the orientation of the clones (centromere–P1 124G1–cDNA 126541–cos 25F11–telomere).
DISCUSSION
The limits of linkage analysis still make positional cloning a difficult task, especially if the patient material is small and none of the patients carry any chromosomal rearrangements to restrict the critical DNA region for a disease gene. Linkage analysis enables at its best the restriction of the region to 1 cM (equal to ∼1 Mb in physical distance). Isolated populations, however, offer a special advantage for fine positioning of disease genes. For example, in Finland, linkage disequilibrium has been utilized in several studies of rare inherited diseases to successfully restrict the DNA region for a disease gene by extrapolating information from meiotic events in previous generations (Hästbacka et al. 1992; Peltonen et al. 1995). Furthermore, linkage disequilibrium analyses allow utilization of the genetic information from affected individuals with no other available samples from the families. Here, we were able to restrict theAPECED region from the original 2.6 Mb to 350 kb by genotyping all existing polymorphisms on the region in 69 APECED families and 30 additional APECED patients with no family data available. Because of the lack of a tight marker map over the region and our unsuccessful efforts to identify novel polymorphisms, we have not been able to maximally utilize linkage disequilibrium in the further restriction of the APECED region. However, potential recombinational events still exist in several families and are to be characterized in the future. One solution to the lack of genetic markers would be the identification of single nucleotide variations that should appear every 1 kb, on average (Kwok et al. 1996). However, the low informativeness of these markers would limit their use in the linkage disequilibrium mapping, unless large numbers of such polymorphisms are to be generated.
The recent progress in enhancing the resolution and sensitivity of the FISH techniques has greatly facilitated the physical map construction (Raap et al. 1995; Heiskanen et al. 1996; van Gijlswijk et al. 1996). In our positional cloning approach on the APECED gene region we have applied FISH methods with different resolution levels. We were able to construct a visual clone contig over an ∼800-kb region covering the APECED gene region on chromosome 21q22.3, including the flanking markers D21S25 andD21S171. The resolution obtained on metaphase chromosomes was not adequate to order different clones spanning the critical region. To obtain a better resolution we applied mechanically stretched chromosomes as targets in FISH, but the method was not applicable on this region. This may be due to the fact that acrocentric and small chromosomes, such as chromosome 21, are not stretched during the procedure as much as large, metacentric chromosomes (Laan et al. 1995). A sufficient resolution level allowing distance measurements and an accurate visual physical map was therefore achieved utilizing the fiber FISH method (Heiskanen et al. 1994).
The fiber FISH method has proved to be equally reliable as other physical map construction methods, such as PFGE, but less time consuming and laborious than traditional mapping methods (Florijn et al. 1995). This method has been utilized previously in the visual physical map construction of several genomic regions linked to human diseases (Heiskanen et al. 1995; Klockars et al. 1996; Leppänen et al. 1996). We applied fiber FISH to localize probes directly in relation to the physical map regardless of the probe size and also irrespective of whether the probes overlap. This enabled us to analyze the order and orientation of the probes as well as their distances from each other. Our data support the earlier observation that the optimal size range for the measurement of clone order and the distances separating different clones is 1–300 kb and occasionally up to 500 kb (Heiskanen et al. 1995). To visualize and localize candidate genes forAPECED we have applied the tyramide-based detection method (Raap et al. 1995). This method facilitated the visualization of small cDNA clones on the APECED region both on metaphase chromosomes and on the physical map by fiber FISH.
The chromosomal regions near centromeres and telomeres of most human chromosomes are known to be problematic for cloning and characterization because of their high content of repetitive and GC-rich sequences (Gardiner 1996). The subtelomeric region of 21q22.3 containing the APECED locus belongs to these chromosomal regions. Despite numerous reports of complete physical maps of chromosome 21 in YAC clones, the 21q22.3 region is poorly represented in the existing YAC libraries and, based on our data, also in multiple types of other long range clone libraries (Chumakov et al. 1992;Nizetic et al. 1994; Korenberg et al. 1995). Parts of theAPECED region have so far been cloned only into cosmids. Both Lafreniere et al. (1995) and Stone et al. (1996) have constructed restriction maps of cosmids including the region between markersD21S25 and D21S154.
Here, we have completed the physical map of the APECEDregion on chromosome 21q22.3 by P1, PAC, and BAC clones. We performed chromosomal walking from the telomeric side of D21S171toward the centromere and identified several P1 and BAC clones containing the previous gap between the DNA markers D21S154and D21S171. In addition, we provide here ∼300 kb extension to the well-characterized bacterial clone contig from marker D21S171 toward the CD18 gene. This part of the contig may assist in the unraveling of the gene defect behind the Knobloch syndrome, the gene of which was recently assigned to the region flanked by markersD21S171 and D21S1446 (Sertie et al. 1996).
The data presented here provide the tools for the final cloning of theAPECED gene, the product of which most likely plays an important role in the development and function of the human immune system. Identification of the defective gene and characterization of the most common APECED mutations in Finland and in the Iranian Jewish population will enable an efficient and exact diagnosis of the disease and may provide new clues to the etiology and pathogenesis of human autoimmune diseases in general.
METHODS
Family Material
The family material consisted of 47 Finnish APECED families and 22 non-Finnish families, a total of 91 affected individuals. In addition to these families, DNA samples from 13 individual Finnish APECED patients and 17 foreign patients (15 of them were from the Iranian Jewish population) were analyzed with tightly linked genetic markers. DNA was extracted from 10–20 ml of peripheral blood samples according to standard protocols (Vandenplas et al. 1984). All the blood samples were drawn in accordance with the Helsinki declaration.
Genotyping
Genotyping, using restriction fragment length polymorphism (RFLP) markers [D21S25/Genome Data Base (GDB) accession no. 158805, D21S154/GDB accession no. 158782, andPFKL/GDB accession no. 156555], was performed by Southern blot hybridization analysis (Southern et al. 1975). Hybridizations were carried out in accordance with standard protocols. Polymorphic microsatellite markers (JAI, D21S1912/GDB accession no. 618065, PFKL/GDB accession no. 179876,D21S171/GDB accession no. 192317, andD21S1903/GDB accession no. 611637) were analyzed using PCR amplification with radioactively labeled primers and size-analyzed by denaturing PAGE as described previously (Aaltonen et al. 1994;Björses et al. 1996).
Linkage Disequilibrium Analyses
The allelic association was calculated according to the formulaP excess = (P affected − P normal)/(1 − P normal), in which P normal equals the frequency of an allele at a marker locus in normal chromosomes, andP affected equals the higher frequency of a certain allele in chromosomes carrying the disease gene. To estimate the location of the APECED gene we analyzed each of the most tightly linked multiallelic loci using the DISLAMB program (v. 1.3). This program applies a likelihood ratio test for linkage disequilibrium, with 1 degree of freedom, irrespective of the number of markers analyzed or the number of alleles at any given locus. Association analysis for multiple loci jointly was performed with the DISMULT program. In this multipoint method, the recombination fraction is fixed between any given map position and each of the marker loci, and the likelihood is maximized, at that map position, over α (proportion of disease alleles originally associated with a certain allele) and n (number of generations since the introduction of the founder disease allele into the population; here,n = 100). This can provide fine control over the magnitude and resolution rate of the observed association, with respect to the genetic distance from the exact location of the disease gene (Terwilliger et al. 1995).
STS Analyses
STS for 21S25, linking clone D21S400, STS for D21S154, and PAC314N7 end fragments (18744 and 23181) were used to screen the genomic libraries. New STSs (23937 and 23938) were also randomly generated from genomic sequence obtained from P1 124G1. The end fragments of BAC 86O23 were directly sequenced through commercial services (STS 86O23.sp6). The sequence information for the STS Z500C was kindly provided by M.-L. Yaspo (pers. comm.). The CD18 primers were obtained from GDB under accession no. 185175. Primer sequences, PCR product sizes, and annealing temperatures for novel STSs are available as on-line supplements at http://www.cshl.org/gr.
Isolation of Genomic Clones
The chromosome 21-specific cosmid library (LL21NCO2 library), P1 (DuPont, Inc., all P1 clones except P1 1226M10), PAC (P. de Jong, Roswell Memorial Institute, Buffalo, NY), and BAC (Genome Systems, Inc. for P1 clone 1226M10 and BAC clones 54L7 and 4D22; Research Genetics, Inc. for all other BAC clones) genomic libraries were screened by PCR with polymorphic markers and STSs from the APECED region. Colony-purified clones were grown overnight in 100 ml of TB (yeast extract, typtone, glycerol in potassium phosphate buffer) with appropriate antibiotic, 50 μg/ml of kanamycin for bacteria-containing cosmid, P1, and PAC clones, and 12.5 μg/ml of chloramphenicol for BAC clones (Shizuya et al. 1992; Ioannou et al. 1994; Shepherd et al. 1994). Cosmid/P1/PAC/BAC DNAs were prepared by alkaline lysis.
Isolation of End Fragments of P1 and PAC Clones
The end fragments of the genomic clones (PAC clones 92C23 and 314N7; P1 clone 579E5) were isolated using the capture PCR method (Lagerström et al. 1991). Fifty nanograms of clone DNA was digested with either AluI, EcoRV, RsaI,PvuII, BglI, or BclII and ligated to two specific linkers in a total volume of 20 μl. The protocol, the linker primer sequences, and the P1-vector derived primers are described in detail elsewhere (Lagerström et al. 1991; Klockars et al. 1996).
ESTs
To identify newly transcribed sequences within theAPECED region, cDNA selection was performed using PAC 92C23 as the genomic template and hybridized with random-primed cDNA from fetal liver, fetal brain, and adult skeletal muscle (Yaspo et al. 1995). The captured cDNAs were annealed to the pAMP10 plasmid system (GIBCO BRL) and hybridized with the PAC 92C23 clone as well as with the cosmids covering the same genomic region. Selected cDNAs were sequenced manually from both ends according to standard protocols. The sequences obtained were systematically analyzed with BLASTn and BLASTx programs using the nonredundant and dbEST databases (Altschul et al. 1990).
FISH
Metaphase Chromosomes as FISH Targets
Peripheral blood lymphocytes were cultured according to standard protocols and to induce a banding pattern cells were treated with 5-bromodeoxyuridine (BrdU) at the early replicating phase (Takahashi et al. 1990; Lemieux et al. 1992). Metaphase slides were pretreated with RNase (100 μg/ml) and pepsin (20 μg/ml), and stained with Hoechst 33258 (1 μg/ml) for 10 min and exposed to UV light (302 nm) for 30 min.
DNA Fibers as FISH Targets
DNA fibers from agarose embedded PFGE blocks spread on a microscope slide were used as a target for FISH (Heiskanen et al. 1994), and a precise physical map was constructed over the region of interest.
Cosmid, P1, PAC, BAC, and cDNA clones were used as probes in this positional cloning approach. Probes were labeled by nick translation either with biotin-11–dUTP (Sigma) or digoxigenin 11–dUTP (Boehringen Mannheim) according to standard protocol.
Hybridizations were carried out in 50% formamide, 10% dextran sulfate in 2× SSC as described earlier (Pinkel et al. 1986; Lichter et al. 1988). Repetitive sequences were suppressed with 10-fold excess of Cot-1-DNA (BRL, Gaithesburg, MD). After overnight incubation, unspecific hybridization signals were eliminated by washing the slides with 50% formamide/2× SSC, twice in 2× SSC and once in 0.5× SSC, at 45°C. Biotinylated probes were detected using TRITC-conjugated avidin D (Vector Laboratories), and FITC-antidigoxigenin antibodies (Sigma Chemicals) were used to visualize digoxigenin-labeled probes. The slides were counterstained with 5 μg/ml of DAPI (4′-6′-diamino-2-phenylindole; Sigma).
Tyramide-Based Detection
To visualize small clones like candidate cDNA clones, a more sensitive biotinylated tyramide-based detection method was applied as described previously (Raap et al. 1995; van Gijlswijk et al. 1996), both on metaphase chromosomes and on DNA fibers.
Digital Image
A multicolor image analysis was used for acquisition, display, and quantification of the hybridization signals of metaphase chromosomes, stretched chromosomes, and DNA fibers. Ten to thirty images from each hybridization were documented. The system contains a Photometrics PXL camera (Photometrics Inc., Tucson, AZ) attached to a PowerMac7100/Av workstation. IPLab software controls were employed for the camera operation, image acquisition, and a Ludl wheel. The measurement of the physical distance over the critical genomic region as well as clone size and interclone distance measurements were performed using IPLab software, and a cosmid clone with known insert size (cos 65F12, 39 kb) was used as a standard for calibration. The distance measurements were confirmed with multiple combinations of different clones.
Acknowledgments
This work was financially supported by the Ulla Hjelt fond of the Foundation for Pediatric Research, the Maud Kuistila Foundation, and the Academy of Finland. The LL21NCO2 and human genomic PAC libraries were the kind gift of Dr. Pieter de Jong. We thank Elina Hellsten and Jouni Vesa for sharing their wide knowledge on positional cloning, Mervi Heiskanen for her expertise on FISH technology, and Anu Wartiovaara for valuable comments on the manuscript. Tuula Airaksinen and Mervi Eeva are appreciated for expert technical assistance. We thank Marion Carson for revision of the English language.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.