Arabidopsis thaliana Centromere Regions: Genetic Map Positions and Repetitive DNA Structure
Abstract
The genetic positions of the five Arabidopsis thalianacentromere regions have been identified by mapping size polymorphisms in the centromeric 180-bp repeat arrays. Structural and genetic analysis indicates that 180-bp repeat arrays of up to 1000 kb are found in the centromere region of each chromosome. The genetic behavior of the centromeric arrays suggests that recombination within the arrays is suppressed. These results indicate that the centromere regions ofA. thaliana resemble human centromeres in size and genomic organization.
Genetic mapping of centromeres is essential for the integration of cytological and genetic maps, and marks an important step toward the molecular characterization of centromeric DNA. Although the centromere is one of the most conspicuous markers on the cytological map, determination of the location of centromeres on genetic maps is frequently difficult to achieve. This is especially true in higher animals and plants where the genetic tools for centromere mapping are limited.
We have been pursuing the characterization of centromere regions of the model angiosperm Arabidopsis thaliana. This plant’s small genome (∼100 Mb/haploid) (Meyerowitz 1994) and relatively low abundance of repetitive DNA [∼10% of total (Leutwiler et al. 1984)] make it well suited for molecular chromosome studies. Aiding in this analysis is the availability of dense genetic maps that exist for each of A. thaliana’s five chromosomes (e.g., Hauge et al. 1993; Lister and Dean 1993). In addition, physical maps are being developed for all A. thaliana chromosomes in the form of overlapping cloned genomic fragments (Schmidt et al. 1995; Zachgo et al. 1996).
Considering the generally advanced molecular characterization of theA. thaliana genome, the genomic organization and genetic location of centromeres in this species remain poorly characterized. The A. thaliana centromere regions are heterochromatic (Schweizer et al. 1987) and contain tandem arrays of related repeats (exhibiting ⩾80% similarity) that are ∼180 bp in length (Martinez-Zapater et al. 1986; Simoens et al. 1988; Maluszynska and Heslop-Harrison 1991), a genomic organization that resembles the ∼170-bp alphoid repeat arrays at primate centromeres (Willard 1990;Pluta et al. 1995). It is not clear whether the 180-bp repeat arrays flank or span A. thaliana centromeres, but data from mammalian systems suggest that the alphoid repeats are intimately associated with the centromere and are likely to play a role in centromere function (Heartlein et al. 1988; Haaf et al. 1992; Tyler-Smith et al. 1993;Brown et al. 1994; Larin et al. 1994; Harrington et al. 1997). A number of other middle-repetitive sequence elements have been found to be associated with the 180-bp repeats in genomic clones (Richards et al. 1991; Schmidt et al. 1995; Pelissier et al. 1996; Thompson et al. 1996a,b), suggesting that islands of more complex sequence arrangement are located in the A. thaliana centromeric regions.
An approximate genetic location of three of the five A. thaliana centromeres (on chromosomes 1, 3, and 5) was achieved by Koornneef and coworkers using telocentric mapping techniques (Koornneef 1983; Koornneef and Van der Veen 1983). More recently, physical mapping projects have localized the centromeres on chromosomes 2 and 4 as gaps in the YAC contig maps that are bordered by genomic clones containing 180-bp repeats (Schmidt et al. 1995; Zachgo et al. 1996). We report here the mapping of all five A. thaliana centromere regions, identified by large polymorphic 180-bp repeat arrays, on the standard recombinant inbred genetic map. The organization and genetic behavior of the 180-bp arrays will also be discussed.
RESULTS
General Organization of Centromeric Arrays of the 180-bpHindIII Repeat
To assess the size and organization of the 180-bp centromeric arrays, we performed genomic Southern analysis using techniques that allow large restriction fragments (up to 2 Mbp) to be examined. We digested megabase-sized A. thaliana genomic DNA with various restriction endonucleases and separated the resulting fragments by pulsed-field gel electrophoresis. Restriction fragments containing the 180-bp repeat were detected by Southern blot hybridization.
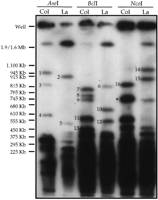
A number of endonucleases that recognize six base-pair sites yielded large (>400 kb), discrete restriction fragments containing 180-bp repeat arrays. We focused our attention on the 180-bp repeat hybridization patterns generated after cleavage with three endonucleases, AseI (ATTAAT), BclI (TGATCA), andNcoI (CCATGG) (see Fig. 1). These endonucleases were chosen in part because they are insensitive to canonical cytosine methylation (mCpG & mCpNpG) (McClelland et al. 1994), which is abundant in the 180-bp repeat arrays (Martinez-Zapater et al. 1986). Although most of the hybridization signals correspond to fragments <400 kb in size, a number of >400-kb restriction fragments were detected. Restriction sites for the three endonucleases are absent from members of the 180-bp repeat family present in the nucleotide databases (data not shown) suggesting that the large restriction fragments are composed of long, uninterrupted arrays of 180-bp repeats or arrays containing a mixture of 180-bp repeats and other low-complexity repeats. We detected only one of the >400-kb fragments seen in Figure 1 after hybridization with probes corresponding to the two other highly reiterated satellite-type repeats abundant in A. thaliana, the so-called 500-bp and 160-bp families (Simoens et al. 1988; Richards et al. 1991) (see Fig. 1), suggesting that most of the large restriction fragments hybridizing with the 180-bp repeat probe are composed primarily of uninterrupted 180-bp repeat arrays.
Identification of large polymorphic restriction fragments containing 180-bp centromeric repeat arrays. High-molecular-weight genomic DNA samples from A. thaliana strain Columbia (Col) or Landsberg erecta (La) were digested with the restriction enzymes indicated and size-fractionated by pulsed-field gel electrophoresis. After electrophoresis, the DNA was transferred to a nylon membrane and hybridized with a radiolabeled 180-bp repeat probe. The polymorphic fragments used for genetic mapping are numbered 1–16 (see Table 1). The fragment indicated by the asterisk cross-hybridized with 160-bp and 500-bp (telomere-similar) repeat probes. The sizes of yeast chromosomal molecular standards are shown at the left.
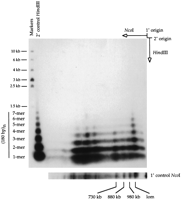
To investigate this point further, we performed a two-dimensional gel analysis to examine the composition of the >400-kb fragments hybridizing to the 180-bp repeat probe. Figure 2demonstrates that the large Landsberg NcoI 180-bp repeat hybridizing fragments can be cleaved completely to small fragments corresponding to the various multimers of the 180-bp repeat seen in the one-dimensional Landsberg/HindIII sample. The variation in the ladder pattern resulting from digestion of different NcoI fragments demonstrates a nonrandom distribution of 180-bp repeat variants.
Two-dimensional gel analysis of large NcoI fragments containing 180-bp centromere repeats. A high-molecular-weight genomic DNA sample from A. thaliana strain Landsberg erecta was digested with NcoI and size-fractionated by pulsed-field gel electrophoresis in the first dimension. An excised gel slice containing the separated NcoI fragments was then incubated withHindIII and placed horizontally at the origin of a 1% agarose gel. After conventional electrophoresis in the second dimension, the fragments were transferred to a nylon membrane and detected by hybridization with a radiolabeled 180-bp repeat probe.HindIII-digested Landsberg erecta genomic DNA was run in the second dimension as a control (shown at left with molecular weight markers). Repeat multimers up to 7-mers were detected. The origin of the second-dimension HindIII control lane was offset slightly from the first-dimension gel slice, accounting for the faster apparent migration of the sample vs. control multimers. A separate first-dimension NcoI control lane is shown at thebottom of the figure. (lom) Limit of mobility.
A significant portion of the A. thaliana genome is accounted for by the long 180-bp arrays. For example, the hybridization patterns seen in Figure 1 indicate that the large (>400 kb) BclI restriction fragments containing 180-bp repeat arrays total over 3 Mbp in length in strain Columbia (see Table 1), corresponding to ∼3% of the ∼100-Mbp haploid A. thaliana genome.
Large Polymorphic Restriction Fragments Containing 180-bp Centromeric Repeat Arrays
Genetic Mapping of A. thaliana Centromere Regions
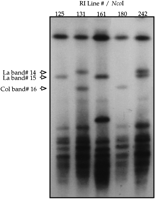
We took advantage of polymorphisms in the size of large 180-bp repeat array restriction fragments to map the genetic location of the five A. thaliana centromere regions (Willard et al. 1986;Kipling et al. 1994). We first identified size polymorphisms in these large fragments between the two common laboratory strains of A. thaliana, Columbia and Landsberg erecta, allowing us to use the Norwich Columbia/Landsberg recombinant inbred (RI) population (Lister and Dean 1993) to map the genetic position of the centromeric repeat arrays. We scored between 29 and 49 independent RI lines for the large polymorphic fragments seen in Figure 1. A total of 16 polymorphic fragments were scored (Table 1; Fig. 3). Five different segregation patterns were discerned, and, in most cases, candidate allelic pairs could be identified. Genotypes for the five patterns were added to the RI mapping database and a genetic map position for each centromere repeat marker, designated RCEN#, was determined by a maximum-likelihood algorithm (Lister and Dean 1993;Arnold and Anderson 1996) (see Fig. 4). The reliability of the RCEN marker locations is reflected by the high logarithim of odds (lod) scores, shown in Table2, which express the relative probability of linkage to a reference marker versus placement at an unlinked location.
Segregation of large polymorphic restriction fragments containing 180-bp centromeric repeat arrays in a recombinant inbred mapping population. High-molecular-weight genomic DNA samples from five A. thaliana Columbia (Col)/Landsberg (La) RI lines were digested withNcoI and size-fractionated by pulsed-field gel electrophoresis. After electrophoresis, the DNA was transferred to a nylon membrane and hybridized with a radiolabeled 180-bp repeat probe. Dominant markers corresponding to fragments 14 and 15 segregated independently. The inheritance of the markers represented by fragments 15 and 16 is mutually exclusive as expected for allelic markers in RI populations (Lister and Dean 1993).
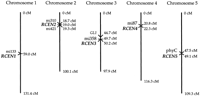
Genetic location of the A. thaliana centromere regions. The locations (in cM) of the centromeric repeat (RCEN) array polymorphisms on the Columbia/Landsberg recombinant inbred map (March 1997 version; http://nasc.nott.ac.uk/) are shown along with relevant reference markers (see text). The mi# markers are as described (Liu et al. 1996). The RCEN markers may be identified on earlier versions of the RI map by their provisional names, EKR#.
lod Scores Assessing the Significance of Linkage of theRCEN Markers to Reference Markers
The 180-bp array polymorphisms map to five positions, one on each of the five A. thaliana chromosomes (Fig. 4) as expected for localized centromere markers. Furthermore, the map positions of the 180-bp array RCEN markers are consistent with the availableA. thaliana centromere mapping information. The map positions of RCEN2 and RCEN4 established here correspond well to the centromere locations deduced from the current physical maps of chromosomes 2 and 4. The RCEN2 marker maps between RFLP markers mi310 and mi421. These markers border gap 1 in the chromosome 2 physical map, which is thought to be the likely position of the centromere (Zachgo et al. 1996). Similarly, the RCEN4 marker is closely linked to the RFLP marker mi87, which is present on candidate centromeric YAC clones containing 180-bp repeats at the end of contig 1 (Schmidt et al. 1995). The map positions of the 180-bp array RCEN1 and RCEN3 markers fall within the genetic intervals containing the centromeres on chromosomes 1 and 3 determined by reference to telocentric chromosome breakpoints. Koornneef and colleagues (Koornneef 1983; Koornneef and Van der Veen 1983) used the telocentric mapping technique to place the centromere of chromosome 1 between the morphological markers tt-1 and ch-1, an ∼5-cM interval that contains the mi133 marker (E.J. Richards, unpubl.). A much larger genetic window for centromere 3, extending ∼40 cM south of gl-1 to tt-5, was defined by the telocentric strategy (Koornneef 1983; Hauge et al. 1993). The 180-bp array RCEN3 marker falls within this interval, ∼5.5 cM south of the GL1 genetic marker. Finally, the telocentric strategy indicated that centromere 5 is located in an ∼11-cM window between morphological markers ga-3 and tt-2(Koornneef 1983; Hauge et al. 1993). Neither of these markers is present on the current recombinant inbred genetic map, yet the localization of the 180-bp array RCEN5 marker in the center of the chromosome is consistent with the position of these morphological markers in the center of the integrated genetic map of A. thaliana chromosome 5. Moreover, RCEN5 corresponds well to the centromere location estimated by the position of repetitive DNA containing clones on the emerging physical map of chromosome 5 (Murata et al. 1997; S. Tabata, pers. comm.; www.kazusa.or.jp/arabi).
DISCUSSION
The identification of large polymorphic restriction fragments containing 180-bp repeats allowed us to determine the genetic locations of the centromeric repeat arrays in A. thaliana. A number of considerations indicate that the mapped centromeric repeats accurately predict the location of the five A. thaliana centromeres. Chromosome in situ hybridization experiments localized the 180-bp repeat exclusively to the primary constrictions and failed to detect interstitial 180-bp repeat loci in the chromosome arms (Maluszynska and Heslop-Harrison 1991). In addition, the 180-bp repeat RFLP markers map to locations consistent with centromere mapping data developed using telocentric approaches or correlations with the gaps in the current physical maps as described in the Results. Moreover, the centromere map positions defined here fall within the genetic intervals containing the meiotic A. thaliana centromeres that have been defined recently by tetrad analysis (G. Copenhaver and D. Preuss, pers. comm.).
Beyond defining useful cytogenetic landmarks on the A. thaliana map, the data presented here provide information on the genomic organization of a centromere from a eukaryotic model organism. The A. thaliana centromere regions are rich in repetitive DNAs, which is a hallmark of the molecularly characterized centromere regions from humans (Willard and Wayne 1987), mice (Kipling et al. 1991; Narayanswami et al. 1992), maize (Alfenito and Birchler 1993;Kaszás and Birchler 1996), Drosophila (Le et al. 1995;Murphy and Karpen 1995), Neurospora (Centola and Carbon 1994), and fission yeast (Nakaseko et al. 1986; Clarke 1990). A. thaliana centromeres contain long arrays of 180-bp repeats with little or no interspersion, in addition to the more complex arrays of 180-bp repeats mixed with other repeat elements characterized previously (Richards et al. 1991; Pelissier et al. 1996; Thompson et al. 1996a,b). A conservative estimate of the size of the uninterrupted 180-bp repeat arrays is ∼0.6 Mbp per centromere assuming an equal distribution of the ∼3 Mbp of uninterrupted 180-bp repeat arrays (summing the BclI fragment lengths; Table 1) among the five centromeres. However, in some cases we can assign >1 Mbp of uninterrupted arrays to a single centromere; for example, centromere 1 (810 + 570 kb in Columbia). Our estimates of the size of theA. thaliana centromere regions are comparable to the size of the alphoid repetitive arrays at human centromeres (Tyler-Smith and Brown 1987; Willard and Wayne 1987; Wevrick and Willard 1989, 1991;Cooper et al. 1993; Jackson et al. 1993).
The size and composition of the 180-bp repeat arrays make it unlikely that a complete A. thaliana centromere can be isolated in a single genomic clone or accurately represented by overlapping clones, using available molecular cloning technology (Neil et al. 1990). The current physical maps of the A. thaliana genome based on overlapping genomic clones do not represent the large 180-bp arrays described here, and we believe that the centromeres are missing from these maps (Schmidt et al. 1995; Zachgo et al. 1996). It may be necessary to analyze A. thaliana centromeres that have been pared down in subchromosomal derivatives rather than rely on assembly of centromeric regions from constituent genomic clones (Tyler-Smith et al. 1993; Brown et al. 1994; Le et al. 1995; Murphy and Karpen 1995;Kaszás and Birchler 1996).
Our analysis of the large polymorphic restriction fragments carrying the 180-bp repeat arrays in the recombinant inbred lines provides insight into the genetic behavior of A. thaliana centromere regions. No nonparental fragments were detected in our analysis, which scored 677 large 180-bp array fragments distributed over 54 RI lines. The absence of nonparental fragments indicates that the centromere repeat arrays are more stable than the long arrays of ribosomal RNA genes [nucleolus organizing regions (NOR)], which have been observed to undergo genomic rearrangements in the recombinant inbred mapping material used here (Copenhaver et al. 1995). Nonparental fragments could also arise from homologous recombination events, including unequal exchanges leading to expansion or contraction of direct repeat arrays, intrachromosomal exchanges leading to deletions, or reciprocal crossover events within the polymorphic arrays. The paucity of homologous recombination events involving the centromeric 180-bp repeat arrays suggests that recombination is suppressed within A. thaliana centromere repeat arrays. Similar conclusions were drawn from the meiotic and mitotic stability of centromeric alphoid repeat arrays in humans (Wevrick and Willard 1989). Recombinational suppression in centromere regions has been observed in other organisms, including mice (Kipling et al. 1994), Neurospora (Centola and Carbon 1994), and fission yeast, Schizosaccharomyces pombe(Nakaseko et al. 1986). The A. thaliana centromere repeats arrays cannot be completely genetically inert, however, as evidenced by the nonrandom distribution of 180-bp variants seen in Figure 2. Similar regional clustering of repeat variants is found in centromeric repetitive DNA arrays in humans (Warburton and Willard 1990) and is thought to reflect intermediates in a recombination-driven homogenization process.
It is clear from our initial characterization of the centromere repeat arrays that A. thaliana centromeres are large, complex regions that resemble their mammalian counterparts. The results described here lay a foundation for further characterization of the centromeres of a model higher eukaryotic organism.
METHODS
Preparation of Large Genomic DNA
Protoplasts from axenic seedling cultures (∼2 weeks old) were prepared and embedded in agarose molds as described (Copenhaver et al. 1995). The embedded protoplasts were lysed and digested at 55°C in two overnight incubations in a solution of 0.5 m EDTA, 1%N-laurylsarcosine, 1 mm Tris at pH 9.5, 1 mg/ml of Pronase (Sigma). The embedded genomic DNA was stored at 4°C in lysis solution minus pronase.
Restriction Endonuclease Digestion and Size Separations
Agarose-embedded DNA samples were treated at 55°C with 100 μg/ml of phenylmethylsulfonyl fluoride for 2 hr to inactivate the protease. The treated samples were then washed in 10 mmTris-Cl, 10 mm EDTA at pH 8.0 before restriction endonuclease digestion. Agarose-embedded DNA samples were digested with 50–100 units of restriction endonuclease (New England Biolabs) in 300 μl of the appropriate buffer + 100 μg/ml of bovine serum albumin + 2.5 mm spermidine. Digested DNA was size-fractionated through a 1% agarose gel (FastLane, FMC) in 0.5× TBE buffer (89 mm Tris-borate, 89 mm boric acid, 2 mm EDTA) using a CHEF (Chu et al. 1986) pulsed-field gel apparatus (CHEF-DRII, Bio-Rad) and the following conditions: 60 sec switch time, 12 hr, 200 V → 90 sec switch time, 9 hr, 200 V at ∼15°C.
Two-Dimensional Gel Electrophoresis
Landsberg genomic DNA was digested with NcoI (New England Biolabs) and size-fractionated on a 1% low-melting point agarose (SeaPlaque, FMC) pulsed-field gel as described above. An excised lane from the first-dimension gel was equilibrated in restriction buffer and subsequently incubated overnight at 37°C after addition of two aliquots of 1000 units of HindIII (New England Biolabs). The gel slice was then equilibrated in 0.5× TBE, cast at the origin of a 1% agarose gel (SeaKem, FMC), and subjected to conventional gel electrophoresis in 0.5× TBE.
Southern Hybridization/Probes
Size-fractionated DNA was depurinated in 0.25 n HCl for 10–15 min, denatured in 0.4 n NaOH, 3 m NaCl and equilibrated in 8 mm NaOH, 3 m NaCl. The nucleic acid was then transferred using a downward capillary apparatus (Scheicher & Schuell) to an uncharged nylon membrane (GeneScreen, DuPont/NEN). After transfer, the membrane was neutralized in 50 mm Na phosphate buffer (pH 6.5) and the nucleic acids were immobilized by UV cross-linking. Hybridizations were carried out in the high-SDS buffer as described (Church and Gilbert 1984). The 180-bp centromere repeat hybridization probe was generated by random-priming DNA synthesis (Ausubel et al. 1987) using the cloned 180-bp repeat monomer from pARR20-1 (E.J. Richards, unpubl.) as a template. The 500-bp repeat hybridization probe was generated by random-priming DNA synthesis using the cloned telomeric insert from pAtT4.1 (Richards and Ausubel 1988; Richards et al. 1991) as a template. The 160-bp repeat hybridization probe was generated by 5′-endlabeling (Ausubel et al. 1987) an oligonucleotide of the following sequence: 5′-TGTTAGTGTTTCTATGGTCA-3′ (Simoens et al. 1988). High-stringency washes in 0.2× SSC, 0.1% SDS at 60°C were used for the 180-bp repeat hybridizations. Even under high-stringency conditions, the 180-bp repeat probe does not exhibit chromosome-specificity, which is characteristic of some human alphoid repeat probes (Willard and Wayne 1987). Lower stringency wash conditions were used to detect the 500-bp repeat (2× SSC, 0.1% SDS at 60°C) and the 160-bp repeat (2× SSC, 0.1% SDS at 45°C). Hybridization signals were detected by autoradiography or PhosphorImager (Molecular Dynamics).
Acknowledgments
This work was supported by a grant from the United States Department of Agriculture (CRG 94-37300-0295). We thank G. Copenhaver for DNA preparations and technical advice. We thank M. Anderson, M. Arnold, C. Dean, and C. Lister for RI mapping resource management (seehttp://nasc.nott.ac.uk/) and the Nottingham Arabidopsis Stock Centre for recombinant inbred stocks.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
We wish to cite recently published cytogenetic analysis of centromeric repetitive DNA organization in A. thaliana byBrandes et al. (1997), as well as previous molecular analysis ofArabidopsis centromeric 180-bp arrays published by Murata et al. (1994).