The structure, function, and evolution of plant centromeres
Abstract
Centromeres are essential regions of eukaryotic chromosomes responsible for the formation of kinetochore complexes, which connect to spindle microtubules during cell division. Notably, although centromeres maintain a conserved function in chromosome segregation, the underlying DNA sequences are diverse both within and between species and are predominantly repetitive in nature. The repeat content of centromeres includes high-copy tandem repeats (satellites), and/or specific families of transposons. The functional region of the centromere is defined by loading of a specific histone 3 variant (CENH3), which nucleates the kinetochore and shows dynamic regulation. In many plants, the centromeres are composed of satellite repeat arrays that are densely DNA methylated and invaded by centrophilic retrotransposons. In some cases, the retrotransposons become the sites of CENH3 loading. We review the structure of plant centromeres, including monocentric, holocentric, and metapolycentric architectures, which vary in the number and distribution of kinetochore attachment sites along chromosomes. We discuss how variation in CENH3 loading can drive genome elimination during early cell divisions of plant embryogenesis. We review how epigenetic state may influence centromere identity and discuss evolutionary models that seek to explain the paradoxically rapid change of centromere sequences observed across species, including the potential roles of recombination. We outline putative modes of selection that could act within the centromeres, as well as the role of repeats in driving cycles of centromere evolution. Although our primary focus is on plant genomes, we draw comparisons with animal and fungal centromeres to derive a eukaryote-wide perspective of centromere structure and function.
During eukaryotic cell division, it is critical that each daughter inherits a balanced chromosome complement, which depends on specialized regions called centromeres (Henikoff et al. 2001; McKinley and Cheeseman 2016). Centromeres function to assemble megadalton protein complexes, called kinetochores, that attach the chromosomes to spindle microtubules and mediate segregation during cell division (Fig. 1A; McKinley and Cheeseman 2016; Musacchio and Desai 2017; Yatskevich et al. 2023). In the majority of eukaryotes, the key determinant for the site of kinetochore assembly is an epigenetic mark: the presence of at least one nucleosome containing a centromere-specific variant of histone 3, called CENH3 in plants and CENPA (also known as CENP-A) in mammals (Fig. 1A; Earnshaw and Rothfield 1985; Talbert et al. 2002; Allshire and Karpen 2008). In humans and yeast, the kinetochore consists of approximately 30 core subunits, within which CENP-A/Cse4 recruits the constitutive centromere-associated network (CCAN) (Yu et al. 2000; Yan et al. 2019; Pesenti et al. 2022; Yatskevich et al. 2022). CCAN forms the inner kinetochore, within which the CENPC and CENPN subunits (also known as CENP-C and CENP-N) directly contact CENP-A (Carroll et al. 2010; Yan et al. 2019; Yatskevich et al. 2022). The CCAN complex also interacts with the Knl1-Mis12-Ndc80 (KMN) outer kinetochore proteins that mediate microtubule attachment (Fig. 1A; Musacchio and Desai 2017; Yatskevich et al. 2023). Knowledge of centromere identity and function is fundamentally important for understanding cell biology and genome architecture across eukaryotes and has applied relevance in the design of stably inherited synthetic chromosomes (Birchler and Swyers 2020; Fachinetti et al. 2020). In plants, centromere-mediated genome elimination also provides a powerful means for haploid induction, which has the potential to accelerate crop improvement (Ravi et al. 2014).
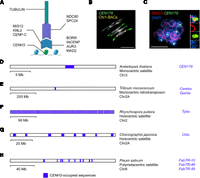
Plant kinetochores and centromere architecture. (A) A diagram of kinetochore structure highlighting known plant proteins, including CENH3 (Talbert et al. 2002), CENP-C (Dawe et al. 1999), MIS12 (Ravi et al. 2011), KNL2 (Lermontova et al. 2013), NDC80 (Du and Dawe 2007), SPC24 (Shin et al. 2018), MAD2 (Yu et al. 1999), AUR3 (Komaki and Schnittger 2017), INCENP (Komaki et al. 2020), and BOREALIN RELATED (BORR) (Komaki et al. 2020). (B) Representative cytological image of the A. thaliana chromosomes during segregation at metaphase (Naish et al. 2021). The DNA is stained with DAPI (white), together with FISH for the CEN178 centromere satellite sequences (green), and Chromosome 1 BAC sequences (yellow) (Naish et al. 2021). Scale bar, 10 μm. (C) Arabidopsis male meiocyte at pachytene stage immunostained for the SMC3 cohesin (red) and stained for DNA (DAPI; blue) (Lambing et al. 2020). FISH was performed against the CEN178 satellite sequence (green). Inset images show magnifications of a CEN178-positive region (Lambing et al. 2020). Scale bar, 10 μm. (D) Representing monocentric satellite architecture, a physical map of A. thaliana Chromosome 3 is shown, with the location of the CENH3-occupied CEN178 array highlighted in blue (Naish et al. 2021). The name of the CENH3-occupied sequences (CEN178) is written alongside in blue. A scale bar is provided indicating physical distance (megabases). (E) Representing monocentric retrotransposon architecture, a physical map of Triticum monococcum Chromosome 2A is shown (Ahmed et al. 2023), as in D. (F) Representing holocentric architecture, a physical map of Rhyncospora pubera Chromosome 2 is shown (Hofstatter et al. 2022), as in D. (G) Representing holocentric architecture, a physical map of Chionographis japonica Chromosome 2A is shown (Kuo et al. 2023), as in D. (H) Representing metapolycentric architecture, a physical map of Pisum sativa Chromosome 6 is shown (Macas et al. 2023), as in D.
Centromeres perform a deeply conserved function across eukaryotes, yet the associated DNA sequences are extremely variable in size and structure, both within and between species (Malik and Henikoff 2009; Rhind et al. 2011; Melters et al. 2013; Logsdon et al. 2023; Wlodzimierz et al. 2023b). For instance, the smallest centromeres, found in budding yeast, are ∼120 bp (Fitzgerald-Hayes et al. 1982; Furuyama and Biggins 2007). These centromere sequences support the loading of a single Cse4 (the CENH3 ortholog) nucleosome and assembly of a single kinetochore complex (Fitzgerald-Hayes et al. 1982; Furuyama and Biggins 2007; Dendooven et al. 2023). In contrast, the centromeres of many plant and animal species consist of megabase tandem repeat arrays that are the site of multiple CENH3 nucleosomes and kinetochore complexes (Fig. 1B–D; Malik and Henikoff 2009; Melters et al. 2013; Naish et al. 2021; Altemose et al. 2022; Wlodzimierz et al. 2023b). A further architectural distinction is seen in holocentric species, in which kinetochores load at multiple sites that are distributed along individual chromosomes (Fig. 1F,G; Heckmann et al. 2013; Drinnenberg et al. 2014; Marques et al. 2015; Schubert et al. 2020; Hofstatter et al. 2022; Kuo et al. 2023), with some species having lost CENH3 or kinetochore proteins altogether (Drinnenberg et al. 2014; Neumann et al. 2023). This contrast between deep functional conservation during chromosome segregation and highly divergent DNA sequences and protein components is termed the centromere paradox (Henikoff et al. 2001; Malik and Henikoff 2009).
In this review, we focus on the structure, function, and evolution of plant centromeres and draw comparisons with animal and fungal genomes to highlight parallel themes and contrasting modes of organization. We review the main architectural types of plant centromeres and their epigenetic states, including patterns of CENH3 nucleosome occupancy and DNA cytosine methylation. Finally, we consider the implications of centromere genetic and epigenetic structure for the observed accelerated rates of evolution within and between species, including the role of recombination.
Monocentric satellite array architecture
Diverse plant species display a monocentric architecture, with a single chromosome constriction observed at metaphase, which contains the kinetochore(s) and attaches to spindle microtubules (Fig. 1A–D; Malik and Henikoff 2009; Melters et al. 2013; Naish et al. 2021). In monocentric plant genomes, the DNA sequences underlying the centromeres are frequently composed of megabase-scale tandem repeat arrays (Malik and Henikoff 2009; Melters et al. 2013; Naish et al. 2021). High-copy tandem repeats are traditionally termed satellites, as following density gradient centrifugation, the repeats form satellite bands owing to different buoyant density compared with the bulk genomic DNA (Kit 1961; Thakur et al. 2021; Altemose 2022). Typically, individual centromeric tandem repeat monomers are between 100 and 200 bp in length and are each capable of hosting a single CENH3 nucleosome (Malik and Henikoff 2009; Melters et al. 2013; Zhang et al. 2013; Maheshwari et al. 2017). Monocentric satellite arrays, which can contain in the range of 1000–10,000 of monomer repeats per chromosome (Fig. 1B,C; Melters et al. 2013; Altemose et al. 2022; Wlodzimierz et al. 2023b), create a DNA substrate with the potential to organize multiple CENH3 nucleosomes and kinetochore complexes (Figs. 1A, 2). This architecture is termed regional organization, in contrast to the point centromeres of budding yeast (Henikoff et al. 2001; Malik and Henikoff 2009).
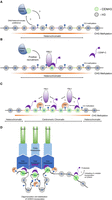
Model for CENH3 loading and centromere homeostasis in Arabidopsis. (A) CENH3 (green circle) is loaded into nucleosomes in a region of CEN178 satellite repeats. The individual repeats are indicated by varying colors. Loading is mediated by a histone chaperone, for example, NASPSIM3 (Le Goff et al. 2020). The region shown is heterochromatic and occupied by nucleosomes containing canonical histone 3 (H3; gray circles). These nucleosomes are modified by H3K9me2, and the associated DNA is methylated in CG, CHG, and CHH sequence contexts. It is possible that heterochromatin marks recruit the CENH3 loading complex. (B) Once CENH3 has been loaded, it assembles kinetochore proteins, including CENP-C and KNL2 (purple) (Ogura et al. 2004; Sandmann et al. 2017; Le Goff et al. 2020). Interactions between the CENH3 loader complex and kinetochore factors may create feed-forward recruitment of CENH3. (C) As the density of CENH3-containing nucleosomes increases, the region undergoes stabilization and compaction of the centromeric chromatin, potentially facilitated by oligomerization of inner kinetochore proteins, for example, CENP-C or KNL2 (Hara et al. 2023; Sissoko et al. 2024). VIM1 (orange) is known to bind and maintain methylation at CG sites and contributes to centromere “strength,” potentially via promotion of kinetochore multimerization. Once a high density of CENH3 nucleosomes is acquired, the region becomes centromeric chromatin, which in Arabidopsis shows reduced CHG context DNA methylation (Naish et al. 2021). (D) Stable loading and maintenance of CENH3 nucleosomes allow recruitment of inner and outer kinetochore complexes (blue) that attach to spindle microtubules (green). We illustrate looping of chromatin via kinetochore multimerization such that CENH3 nucleosomes are gathered in space. Outside of this region, CENH3 and kinetochore proteins are unstable owing to the action of removal pathways, potentially including SUMO or ubiquitin-mediated proteolysis.
Our understanding of centromere tandem repeat arrays has been limited in many species, as they could not be fully assembled using short-read sequencing (Miga and Sullivan 2021). However, Oxford Nanopore Technology (ONT) and Pacific Biosciences (PacBio) HiFi long-read sequencing now allow complete assembly of complex centromere-associated tandem repeat arrays in plants and animals (Miga et al. 2020; Naish et al. 2021; Altemose et al. 2022; Nurk et al. 2022; Logsdon et al. 2023; Wlodzimierz et al. 2023b). Representative assembled plant genomes with monocentric tandem repeat architecture include Arabidopsis thaliana (CEN178 satellites are ∼178 bp) (Naish et al. 2021; Wlodzimierz et al. 2023b), Arabidopsis lyrata (CEN168 and CEN179 satellites are ∼168 and ∼179 bp) (Wlodzimierz et al. 2023b), Brassica rapa (CentBr satellites are ∼176 bp) (Zhang et al. 2023a), Vitis vinifera (satellites are ∼107 bp on 16 of 19 chromosomes) (Shi et al. 2023), Oryza species (CentO satellites are ∼155 bp) (Cheng et al. 2002; Song et al. 2021), Vigna unguiculata (CEN455, CEN721, CEN1600 satellites are 455, 721 and 1600 bp, respectively) (Yang et al. 2023), Glycine max (GmCent satellites are 92 bp) (Liu et al. 2023), and Erianthus rufipilus (CEN137 satellites are 137 bp) (Fig. 1D; Table 1; Wang et al. 2023b). Atypically long (up to 5-kb) and short (44-bp) satellite repeats have also been reported in potato and Vicia faba, respectively (Gong et al. 2012; Ávila Robledillo et al. 2018). As complete genome sequences accumulate, methods to de novo identify and classify satellite repeats, for example, TRASH, SRF, and TRF, will be important for centromere analysis (Benson 1999; Wlodzimierz et al. 2023a; Zhang et al. 2023b).
Plant genomes that represent different centromere architectural types
In monocentric species, centromeric satellite arrays are typically localized to a single chromosomal region. However, during evolution, novel arrays can emerge, expand, and replace existing repeats, or arrays may be split through chromosome rearrangements, leading to multiple distinct satellite regions on the chromosome (Liu et al. 2023; Logsdon et al. 2023; Wlodzimierz et al. 2023b). In chromosomes with distinct satellite arrays, frequently only a single array, or subset of repeats, will be CENH3-occupied (Naish et al. 2021; Song et al. 2021; Chen et al. 2023; Wlodzimierz et al. 2023b). For example, in A. thaliana, considerable variation in CEN178 array size and structure was observed across a sample of four accessions (1.5–6.5 Mb), yet in all cases a ∼1- to 2-Mb region of satellite repeats was occupied by CENH3 (Wlodzimierz et al. 2023b). In contrast to A. thaliana, where the centromeres are composed of the same CEN178 repeat family, in the sister species A. lyrata four centromeres consist of the CEN179 repeat and four centromeres of the CEN168 repeat (Kawabe and Nasuda 2005; Berr et al. 2006; Lysak et al. 2006; Wlodzimierz et al. 2023b). Furthermore, other species have a more heterogeneous centromere satellite organization. For example, interspecific hybrid aspen (Populus tremula × P. alba), maize (Zea mays), and potato (Solanum tuberosum) possess centromeres with and without satellite tandem repeat arrays (Gong et al. 2012; Bao et al. 2022; Chen et al. 2023; Zhou et al. 2023). Together, these patterns are consistent with rapid turnover and dynamic evolution of satellite repeats in plant species.
High levels of satellite repeat sequence polymorphism have been observed within plant genomes. For example, in A. thaliana, numerous CEN178 satellite polymorphisms exist, many of which are private to individual chromosomes (Naish et al. 2021; Wlodzimierz et al. 2023b). A similar pattern occurs in rice and Erianthus, in which satellites are more similar within chromosomes than between (Song et al. 2021; Wang et al. 2023b). These patterns indicate that mutations arising within different centromeres rarely spread to the other chromosomes, implying that inter-chromosome centromere recombination or repair is infrequent, at least within these species. Satellite monomers are also commonly arranged in higher-order repeats (i.e., monomer satellites a, b, and c in an abcabcabc arrangement, where abc is the higher-order repeat), and these arrangements can also be highly polymorphic (Naish et al. 2021; Altemose et al. 2022; Logsdon et al. 2023; Wang et al. 2023b; Wlodzimierz et al. 2023b). In A. thaliana, CEN178 higher-order repeats commonly involve two or three monomers that occur in close proximity (<10 kb) (Naish et al. 2021; Wlodzimierz et al. 2023b). Nonetheless, there are also instances in which large duplications spanning >100 kb in length are observed (Naish et al. 2021; Wlodzimierz et al. 2023b). In both Arabidopsis and Erianthus, higher-order duplications are often separated by megabase distances, consistent with long-range recombination (Naish et al. 2021; Wang et al. 2023b; Wlodzimierz et al. 2023b). In humans, alpha-satellite HOR patterns are more regimented and homogenized compared with Arabidopsis and involve a greater number of monomers per HOR (between four and 19) (Rudd et al. 2006; Altemose et al. 2022; Wlodzimierz et al. 2023a). Notably, each human chromosome is typified by specific HOR patterns (Rudd et al. 2006; Altemose et al. 2022), which is further consistent with satellite recombination being generally restricted within chromosomes, as in Arabidopsis, rice, and Erianthus.
As further complete genomes accumulate, it will be possible to analyze pancentromeric diversity within populations. For example, analysis of satellite arrays within 66 A. thaliana accessions has revealed high levels of structural diversity, although the same CEN178 family of repeats composed all centromeres (Wlodzimierz et al. 2023b). Currently, mutation rates in plant centromeres have not been reported, but evidence in human centromeres and Arabidopsis pericentromeric heterochromatin indicate elevated rates compared with the chromosome arms (Weng et al. 2019; Logsdon et al. 2023). Evidence for lateral spread of mutations between satellite sequences within the same centromere is widespread (Dover 1982; Rudd et al. 2006; Wlodzimierz et al. 2023b). Analysis of variants within the Arabidopsis satellites showed signatures of sequence homogenization, reminiscent of concerted evolution in other tandem repeat arrays (Coen et al. 1982; Durfy and Willard 1990; Liao et al. 1997), where high frequency CEN178 variants are found in the center, rather than the edges, of the centromeric arrays (Wlodzimierz et al. 2023b). Concerted evolution of satellites implies that an active process of DNA breakage and homologous recombination occurs within the center of the arrays, and indirect evidence for this exists in multiple species, including humans and maize (Chatterjee and Lo 1989; Mahtani and Willard 1990; Nijman and Lenstra 2001; Rudd et al. 2006; Shi et al. 2010; Wolfgruber et al. 2016). Centromeric satellite recombination could occur during meiosis, in which either a homolog or a sister chromatid may be used for repair of SPO11-induced breaks. Alternatively, recombination could occur during mitosis, in which sister chromatid recombination is most likely, as interhomolog recombination is typically suppressed. Intra-chromatid recombination between linked loci is also possible, and consistent with this, DNA breaks in human centromeres during the G1 phase of the cell cycle are actively repaired using RAD51, despite the absence of a replicated sister chromatid (Yilmaz et al. 2021).
Because of the observed high levels of satellite structural polymorphism, it has been proposed that unequal crossover could contribute to centromere evolution (Smith 1976). In this influential model, meiotic DNA double-strand breaks (DSBs) are repaired using a nonallelic location on a homolog, with the potential to cause gain and loss of intervening tandem repeats (Smith 1976). However, it is widely documented that centromeres and flanking sequences are potently suppressed for meiotic crossover across eukaryotes (Mahtani and Willard 1998; Vincenten et al. 2015; Hartmann et al. 2019; Naish et al. 2021; Fernandes et al. 2024), meaning unequal interhomolog crossover is unlikely as a general mechanism for centromere evolution. For example, in A. thaliana, haplotype linkage blocks have been maintained across similar CEN178 arrays, despite internal satellite dynamics, where array expansion and shrinkage has occurred since populations diverged (Wlodzimierz et al. 2023b). This is similar to maintenance of large centromere-spanning haplotypes, termed cenhaps, observed in humans (Langley et al. 2019), within which satellite arrays also evolve dynamically (Logsdon et al. 2023). Together, these patterns are consistent with unidirectional allelic or nonallelic gene conversion mediating array evolution, or unequal crossover between sister chromatids, rather than interhomolog unequal crossover. Further support for conversion-type pathways being a dominant mode of recombination within the centromeres was seen in maize, in which CRM retrotransposons were eliminated during meiosis, without exchange of flanking markers (Shi et al. 2010).
A further pathway with the potential to mediate change to centromere satellite arrays is break-induced replication (BIR) (Rice 2020; Haig 2022; Talbert and Henikoff 2022; Showman et al. 2023). BIR can repair DNA DSBs via one of the broken ends being resected, allowing it to invade another chromosome, where it acts as a primer for DNA synthesis (Liu and Malkova 2022). Following initiation of replication, unidirectional migration of a replication bubble may occur over hundreds of kilobases, followed by DNA repair (Liu and Malkova 2022). In centromere repeat arrays, there may be multiple locations to which the invading end can anneal and prime replication, with the potential to cause deletions, duplications, or conversion of satellite repeats (Liu and Malkova 2022; Talbert and Henikoff 2022). Centromeric DSBs may form via tension during chromosome segregation or owing to stalled DNA replication on repetitive templates (Scelfo and Fachinetti 2023). Support for the BIR model comes from recent work quantifying alpha-satellite repeat array size over about 20 mitotic cell divisions of human cell lines (Showman et al. 2023). This revealed significant copy number change over this time course and revealed RAD52 and PIF1 helicase mediate these dynamics, which is consistent with BIR (Showman et al. 2023).
It has been speculated that recombination factors that are directly associated with CENP-A/CENH3 and/or the kinetochore could promote instability of satellite repeats, termed the kinetochore associated recombination machine (KARM), or KARM in Arabidopsis (KARMA) (Fig. 3C; Miga and Alexandrov 2021; Wlodzimierz et al. 2023b). Consistent with this model, purified meiotic kinetochores from budding yeast contain recombination factors, including Mer2, Rad51, Rad59, Spo11, and Spp1 (Borek et al. 2021). Further, proteomic analysis of human alpha-satellite DNA in Xenopus egg extracts also identified multiple DNA damage and repair proteins, including MRE11-RAD50, PARP1, Ku80, MSH2-MSH6m XRCC1, XRCC5, and MUS81 (Aze et al. 2016; Scelfo and Fachinetti 2023). However, the precise recombination factors and pathways that are associated with plant kinetochores remain unknown. The KARMA model makes specific predictions, including that CENH3 enrichment should correlate with the most homogenous repeat arrays, which has yet to be fully tested. It is also worth noting that other tandem repeat arrays, for example, rDNA, show similar patterns of concerted evolution, yet lack CENH3 and kinetochore occupancy (Coen et al. 1982; Liao et al. 1997), and therefore, additional pathways for sequence homogenization must also exist.
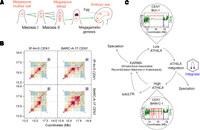
Centromere drive and evolution in plants. (A) Diagram showing differentiation of the megaspore mother cell (MMC) in a subapical position of an Arabidopsis ovule (red). The MMC undergoes meiosis resulting in four haploid daughter cells, three of which enter programmed cell death, leaving a functional megaspore that differentiates into the haploid embryo sac. The embryo sac contains the egg cell, which is fertilized by pollen to produce progeny seed. The green arrows indicate the theoretical path a selfish centromere could follow to overtransmit itself in a non-Mendelian manner, causing meiotic drive. (B) StainedGlass sequence identity heat maps comparing CEN1 within and between IP-Ini-0 (Spanish) and BARC-A-17 (French) accessions of A. thaliana (Vollger et al. 2022; Wlodzimierz et al. 2023b). Red and orange indicate high levels of sequence similarity. The gray lines demarcate a ∼1-Mb BARC-A-17 region that is similar to a ∼100-kb IP-Ini-0 region, indicating centromere sequence dynamics since these accessions diverged (Wlodzimierz et al. 2023b). (C) A cyclical model for centromere evolution in A. thaliana, which we propose alternates between high and low ATHILA retrotransposon invasion states, represented by CEN1 in the Bon-1 and BANI-C-1 accessions respectively, which belong to the same satellite similarity group, yet differ in the level of ATHILA invasion (Wlodzimierz et al. 2023b). ATHILA transcription generates new copies with the potential to integrate into the centromeres. Intact ATHILA may undergo internal recombination to generate soloLTRs. Simultaneously, satellite arrays undergo recombination, resulting in repeat homogenization and purging of ATHILA via a proposed kinetochore associated recombination machine in Arabidopsis (KARMA) (Miga and Alexandrov 2021; Wlodzimierz et al. 2023b). Inter-species satellite and retrotransposon polymorphisms are also consistent with roles of centromere evolution in speciation.
CENH3 homeostasis within plant centromeres
Structural studies have revealed the integral role of CENP-A nucleosomes in contacting other mammalian kinetochore proteins, including the CCAN subunits CENP-C and CENP-N (Yan et al. 2019; Pesenti et al. 2022; Yatskevich et al. 2022). Thus, central to understanding plant centromere identity are the mechanisms by which CENH3 nucleosomes are deposited de novo (established), maintained through DNA replication and cell division, and unloaded from the chromosome or degraded (Fig. 2). It is important to note that CENH3 ChIP-seq of cell populations analyses the steady state of enrichment, which represents an equilibrium between establishment, maintenance, and removal processes. In this section, we consider what is known about these pathways in plants and how this relates to dynamic and homeostatic patterns of CENH3 occupancy.
Histones are loaded into chromatin by a wide range of chaperone proteins (Fig. 2A; Yadav et al. 2018). In animals, HJURP is a major CENP-A chaperone, which functions during the G1 cell cycle phase (Dunleavy et al. 2009; Foltz et al. 2009). Tethering HJURP with the LacI repressor to a LacO array is sufficient to drive ectopic CENP-A loading and kinetochore formation in human cells, which was dependent on the Mis18 kinetochore protein (Barnhart et al. 2011). This is consistent with interaction studies that show HJURP directly binds both Mis18 and CENP-A:H4 tetramers (Pan et al. 2019). This indicates a reinforcement relationship, in which Mis18 recruits HJURP, which loads CENP-A, which would then recruit further Mis18 (Barnhart et al. 2011). In fungi, Scm3 functions as the Cse4/Cnp1 loading chaperone in budding and fission yeast, which is distantly related to HJURP (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007; Pidoux et al. 2009; Sanchez-Pulido et al. 2009; Williams et al. 2009; Shivaraju et al. 2011). Further loading factors have been identified, including CAL1 that loads Cid/CenH3 in Drosophila (Chen et al. 2014), and NUCLEAR AUTOANTIGENIC SPERM PROTEIN (NASP), which functions as a Cnp1 loader in fission yeast (Dunleavy et al. 2007). To date, no orthologs of HJURP/Scm3 or CAL1 have been identified in plants, although an Arabidopsis ortholog of NASP/SIM3 binds to CENH3 in vitro, and RNAi lines cause reduced CENH3 loading in vivo (Le Goff et al. 2020). In A. thaliana, mutation of an KMN complex ortholog, KNL2, shows reduced CENH3 loading and cell division defects (Lermontova et al. 2013), which is similar to KNL1/knl-1 mutants in humans and Caenorhabditis elegans, respectively (Hayashi et al. 2004; Fujita et al. 2007; Maddox et al. 2007). The Arabidopsis gamma-tubulin complex protein3–interacting protein double-mutant gip1 gip2 also shows decreased CENH3 loading and defects in centromeric cohesion (Batzenschlager et al. 2015). These findings are generally consistent with a positive feedback loop between kinetochore proteins and CENH3 acting in plants, potentially via recruitment of loading factors analogous to HJURP or via stabilization of neighboring CENH3 incorporation (Fig. 2B). It will be important to identify mechanisms of CENH3 loading in plant genomes and how the factors involved are themselves recruited to the chromosomes.
In addition to interacting chaperones, different regions of CENH3 itself are known to be required for specific loading patterns into chromatin. CENH3 possesses a highly conserved C-terminal histone fold, as well as a variable N-terminal tail that is diverged from canonical histone 3 (Talbert et al. 2002). In Arabidopsis, the C-terminal CENH3 histone fold domain, including the variable loop 1 region, is sufficient to direct centromere targeting, even when the N-terminal tail is absent (Lermontova et al. 2006). Further work in Arabidopsis has shown that mutation of a conserved lysine within the C-terminal domain at position 130 (L → I or L → F) is sufficient to impair the centromeric deposition of CENH3 (Karimi-Ashtiyani et al. 2015). Interestingly, CENH3-tailswap-GFP fusion proteins with intact C-terminal domains show loading onto chromosomes during mitosis but not meiosis, which implies differential control between these cell types mediated via N-terminal tail interactions (Ingouff et al. 2007; Ravi et al. 2011). Recent work has shown that oligomerization of mammalian inner kinetochore proteins, such as CENP-C (Hara et al. 2023) and CENPT (also known as CENP-T) (Sissoko et al. 2024), can stabilize and protect CENP-A loading and promote kinetochore assembly. As CENP-C is conserved in plants (Ogura et al. 2004; Du and Dawe 2007), similar oligomerization mechanisms may operate during plant kinetochore formation (Fig. 2C,D).
To investigate control of centromere identity, an A. thaliana cenh3-null mutant was transformed with Z. mays or Lepidium oleraceum CENH3 transgenes (Maheshwari et al. 2017). Despite the CENH3 proteins being divergent and adapted to unrelated centromere sequences, both variants remarkably localized to the A. thaliana CEN178 satellite arrays (Maheshwari et al. 2017). This implies that centromere epigenetic or topological states, rather than the primary satellite sequence, direct CENH3 loading. Although the mechanism of CENH3 loading in plants remains unclear, evidence suggests that CENH3 occupancy is dynamic and homeostatic. For example, transfer of maize chromosomes into oat (Avena sativa), via interspecies crossing, showed that regions of CENH3 ChIP-seq enrichment expanded from ∼1.8 Mb to ∼3.6 Mb, once in an oat nucleus (Jin et al. 2004; Wang et al. 2014). This indicates that the nuclear context, and likely trans-acting factors, can influence CENH3 occupancy on centromeric DNA sequences. In Arabidopsis, most CENH3 loading occurs during the G2 phase of the cell cycle (Lermontova et al. 2006), which may contribute to a homeostatic limit for centromere size. Further consistent with this idea, Arabidopsis cells with increasing ploidy show a linear increase in satellite FISH signal but without a proportional increase in CENH3 immunostaining (Lermontova et al. 2006).
Dynamic centromere positioning has been observed via cytogenetic mapping within Arabideae, soybean, and cucurbit species, which is thought to play a role in reproductive isolation and speciation (Han et al. 2009; Mandáková et al. 2020; Liu et al. 2023). In these species, the centromeres were observed to migrate to form evolutionary new centromeres (ENCs), without large-scale chromosome rearrangements (Han et al. 2009; Mandáková et al. 2020; Liu et al. 2023), which parallels similar observations of ENCs made in mammals (Carbone et al. 2006). For example, the region of CENP-A enrichment on horse Chromosome 11, which lacks satellite DNA, is found to migrate within a ∼500-kb range in different individuals but remains static within different tissues of the same individual (Cappelletti et al. 2023). Dynamic localization of CENH3 has also been observed on maize and wheat chromosomes during inbreeding (Benson 1999; Wlodzimierz et al. 2023a; Zhang et al. 2023b; Zhao et al. 2023) and observed when comparing multiple soybean genomes (Liu et al. 2023). Additionally, wheat CENH3 has been shown to preferentially associate with rye repeat satellites following chromosome fusion, rather than the native centromere repeats (Karimi-Ashtiyani et al. 2021). These studies show epigenetic migration of the centromere to new DNA sequences over short timescales. However, the factors that dictate the rate and extent of centromere migration during inbreeding and ENC formation are not completely understood.
In humans, deletion of the endogenous centromere arrays can trigger CENP-A loading and neocentromere formation in new locations (Marshall et al. 2008; Scott and Sullivan 2014). For example, chromosome engineering was used to delete alpha-satellite arrays in human cells, which triggered formation of a CENP-A neocentromere in a ∼100-kb gene-poor domain associated with elevated levels of the heterochromatic mark H3K9me3 (Murillo-Pineda et al. 2021). In maize, UV-irradiation produced a Chromosome 3 fragment (Dp3A) that showed CENH3 loading on gene-associated sequences that are not occupied on the intact chromosome (Fu et al. 2013). Stably maintained deletion derivatives of barley chromosomes have also been obtained, in which the endogenous centromere repeats have been deleted and neocentromeres created (Nasuda et al. 2005). This indicates a “pressure” to load a focus of CENH3, such that if the preferred locations are removed or epigenetic memory is lost, alternative loci can assume centromere identity. Although this suggests significant flexibility in CENH3 loading, it is also clear that there are likely strong effects of DNA sequence. For example, CENH3 has been mapped in different A. thaliana strains (Col, Ler, Cvi, and Tanz) with varying CEN178 arrays. In a subset of cases, CENH3 was found to occupy CEN178 subarrays that are defined by specific sequence variants, consistent with preferential loading at the DNA level (Wlodzimierz et al. 2023b). As commented on earlier, across the varying Arabidopsis CEN178 array sizes tested, CENH3 enrichment occurs in a consistent ∼1- to 2-megabase region in each case, which implies a homeostatic limit that prevents full occupancy of the available satellite arrays (Wlodzimierz et al. 2023b). In contrast, despite human alpha-satellite arrays spanning a similar multi-megabase scale, a much smaller ∼10- to 20-kb region in each array shows CENP-A enrichment (Altemose et al. 2022; Logsdon et al. 2023), indicating a potentially stronger homeostatic limit on centromere identity in humans.
Although native CENH3/CENP-A is loaded during specific phases of the cell cycle, transient or ectopic expression has been performed in plants and animals to investigate control of deposition into chromatin (Heun et al. 2006; Moreno-Moreno et al. 2006; Feng et al. 2020). For example, in maize, a CENH3-GFP transgene driving more than 100-fold higher expression than the wild type, showed only a modest increase in CENH3 loading into the centromere, indicating limits on deposition (Feng et al. 2020). In Drosophila, inducible overexpression of the centromeric histone CID resulted in broad mislocalization along the chromosomes associated with cell lethality and lagging or fragmented chromosomes (Heun et al. 2006). Interestingly, transfection of a CID-YFP expressing plasmid in Drosophila Kc cells initially showed broad mislocalization, yet as the cell culture progressed, CID-YFP became limited to the native centromeres, suggesting dynamic and ongoing regulation (Heun et al. 2006). In yeast and Drosophila, ubiquitin-proteasome-mediated proteolysis plays important roles in limiting and removing ectopic euchromatic Cse4/CID (Collins et al. 2004; Moreno-Moreno et al. 2006). Similarly, in humans, inner kinetochore proteins become densely SUMOlyated in a senp6 SUMO-protease mutant, which causes them to be degraded via p97 (Mitra et al. 2020; van den Berg et al. 2023). Although CENH3 overexpression has not been reported in Arabidopsis, the cenh3-4 splice acceptor allele shows a 10-fold reduction of CENH3 mRNA and significantly reduced protein accumulation (Capitao et al. 2021). Despite this, cenh3-4 plants are viable, are fertile, and do not show obvious growth phenotypes, although there are defects in chromosome stability following heat stress (Capitao et al. 2021; Ahmadli et al. 2023). It will be interesting to understand how the processes that control loading and removal of plant CENH3 adapt to low protein abundance backgrounds such as cenh3-4, as well as to overexpression.
CENH3, genome elimination, and mini-chromosomes
Genetic experiments that manipulated CENH3 in Arabidopsis, maize, and wheat have triggered genome elimination, when chromosomes carrying variant CENH3 segregate with wild-type chromosomes during embryonic development (Ravi and Chan 2010; Karimi-Ashtiyani et al. 2015; Kuppu et al. 2015; Tan et al. 2015; Lv et al. 2020; Wang et al. 2021). As a consequence, altering CENH3 can act as an efficient trigger of uniparental genome elimination and haploid induction. This shows both the dramatic effect that changes to the centromere can have on genome transmission, in addition to providing a powerful tool during breeding and genetic analysis (Ravi and Chan 2010; Ravi et al. 2014; Lv et al. 2020; Wang et al. 2021).
Early work in A. thaliana showed that null cenh3 mutations, which are lethal, could be partially complemented by modified CENH3 transgenes, including a version with the histone tail replaced with that from histone 3.3, and additionally fused to GFP (CENH3-tailswap-GFP) (Ravi and Chan 2010). When CENH3-tailswap-GFP lines were crossed to the wild type, a high proportion (25%–45%) of the progeny were haploid and had lost the chromosomes from the CENH3-tailswap-GFP parent (Ravi and Chan 2010). Genome elimination occurs in early embryonic cell divisions and is associated with chromosome missegregation, formation of micronuclei, and rearranged chromosomes (Tan et al. 2015, 2023; Marimuthu et al. 2021). This phenomenon is reminiscent of chromothripsis and chromoanagenesis, in which DNA breakage leads to chromosome rearrangements (Ly and Cleveland 2017; Guo et al. 2023). During haploid induction, centromere dysfunction may lead to inappropriate positioning of chromosomes during cell division and breakage owing to shearing or mechanical stresses. Genome elimination has been achieved by crossing lines carrying point CENH3 mutations in Arabidopsis (Karimi-Ashtiyani et al. 2015; Kuppu et al. 2015), by genome-editing to produce frameshifts in CENH3 tail regions in wheat (Lv et al. 2020), and by crossing maize cenh3-null heterozygotes (Wang et al. 2021). The consensus model for these effects is that genetic changes to CENH3 that weaken centromere identity during plant gametophytic divisions cause genome elimination when these chromosomes compete with the wild type during embryonic mitosis.
Recent characterization of haploids produced from CENH3-mediated genome elimination has revealed the presence of mini-chromosomes that include the endogenous centromeres, which can be maintained through multiple subsequent generations (Tan et al. 2023). Mini-chromosomes in plants can be linear with telomere caps or with circular rings that are covalently joined (Birchler and Swyers 2020). Analysis of B Chromosome-derived mini-chromosomes in maize showed that many can also pair and recombine during meiosis, although smaller variants showed precocious separation during anaphase and, in other cases, did not pair (Han et al. 2008). In maize, tethering CENH3 using a LexA fusion to an array of LexO repeats causes formation of dicentric chromosomes that show instability and chromosome breakage, with the resulting mini-chromosomes being maintained through subsequent generations (Dawe et al. 2023). As these mini-chromosomes can autonomously propagate, they have the potential to form chassis or blueprints for artificial chromosomes capable of delivering complex pathways within and between species (Dawe et al. 2023). Equivalent experiments that tethered CENH3 to LacO arrays in Arabidopsis were sufficient to drive kinetochore assembly and formation of anaphase bridges (Teo et al. 2013). Analysis of maize plants containing B–A translocations, in which breakage–fusion–bridge cycles occur, has revealed chromosomes carrying two regions with DNA sequences typical of centromeres (Han et al. 2006). These fusion chromosomes showed a single CENH3 focus despite being dicentric at the sequence level, indicating epigenetic inactivation of one centromere (Han et al. 2006). Further studies performed X-ray irradiation of maize tassels carrying supernumerary B chromosomes and screened for chromosomal variations using FISH (Liu et al. 2020). This study identified dicentric chromosomes, only some of which showed a single primary constriction, further consistent with centromere inactivation, in addition to acentric chromosome fragments that established CENH3 foci de novo (Liu et al. 2020). Similarly, engineering of human dicentric chromosomes lead to a variety of consequences, including centromere inactivation (Higgins et al. 2005). Together, these studies indicate that chromosome rearrangements can be associated with the rapid birth and death of centromeres in plants and animals, further implying dynamic and homeostatic mechanisms of CENH3/CENP-A loading and centromere identity.
Centrophilic retrotransposons in plant genomes
Plant centromeres are frequently associated with long terminal-repeat (LTR) class retrotransposons, with some subfamilies termed centrophilic (Nagaki et al. 2005b; Sharma and Presting 2014; Wang et al. 2023b; Naish et al. 2021; Ahmed et al. 2023; Wlodzimierz et al. 2023b). In A. thaliana, centrophilic ATHILA retrotransposons have integrated into the CEN178 repeat arrays and show relatively low CENH3 occupancy compared with the surrounding satellite repeats (Naish et al. 2021; Wlodzimierz et al. 2023b), whereas in other genomes, retrotransposons correspond to the CENH3-occupied sequences themselves (Fig. 1E; Table 1). For example, monocot centromeres frequently contain a high frequency of Gypsy-LTR chromovirus retrotransposons, including CRM in maize, CRR in rice, and Cereba and Quinta in wheat, which can be CENH3-occupied (Presting et al. 1998; Nagaki et al. 2005b; Gao et al. 2008; Neumann et al. 2011; Li et al. 2013; Sharma and Presting 2014). CRM family retrotransposons are typified by a chromodomain fusion at the C terminus of the integrase open reading frame (Gao et al. 2008). As integrase proteins govern LTR retrotransposon and retroviral target site integration (Abascal-Palacios et al. 2021; Maertens et al. 2022), and chromodomains are known to bind histones with specific methylation states (Jacobs and Khorasanizadeh 2002), it is proposed that the integrase–chromodomain fusions target retrotransposon insertion into the centromeres. For example, the chromodomain may directly bind CENH3, kinetochore proteins, or other centromeric chromatin marks to guide the transposon synaptic complex during integration. However, in other plants, for example, Arabidopsis, the centromeric satellite arrays are invaded by ATHILA, which are nonchromovirus LTR retrotransposons that lack integrase-chromodomain fusions (Naish et al. 2021; Wlodzimierz et al. 2023b), indicating that there are likely independent mechanisms of centrophilic adaptation. Furthermore, in A. lyrata, a sister species which is 5 million years diverged from A. thaliana, Copia family LTR retrotransposons, including Tal1, are found within the satellite repeat centromere arrays, in addition to ATHILA (Tsukahara et al. 2012; Wlodzimierz et al. 2023b), and Ale COPIA elements have colonized the centromeres of Brassica nigra (Perumal et al. 2020). As Gypsy and Copia retrotransposon families diverged ∼1 billion years ago (Llorens et al. 2009), this is further consistent with multiple LTR families convergently evolving centrophilic adaptations in different plant lineages.
In the case of Arabidopsis, the majority (>90%) of the centromeres are composed of CEN178 satellite repeats, with a low level (∼1%–10%) of ATHILA invasion (Naish et al. 2021; Wlodzimierz et al. 2023b). However, in rice and maize, although some regions of CENH3 enrichment overlap satellite repeats, many regions are instead dominated by chromovirus CRM and CRR insertions, respectively (Cheng et al. 2002; Zhong et al. 2002; Nagaki et al. 2005b; Song et al. 2021; Chen et al. 2023). In other plants, the centromeric regions of CENH3 occupancy are entirely composed of retrotransposons. A notable case is Einkorn wheat, in which the CENH3-enriched metacentric regions (∼4–5.8 Mb) show an absence of tandem repeat arrays, and instead, 91%–97% of the sequence is composed of LTR retrotransposons of the Cereba and Quinta chromovirus families (Fig. 1E; Table 1; Liu et al. 2008; Li et al. 2013; Ahmed et al. 2023). The Einkorn wheat centromeres are observed to contain the youngest copies of Cereba and Quinta (Liu et al. 2008; Li et al. 2013; Ahmed et al. 2023), which parallels observations of ATHILA in A. thaliana, in which the youngest copies were observed within the core of the satellite repeat arrays (Naish et al. 2021; Wlodzimierz et al. 2023b). Additional examples of retrotransposon-based centromere architecture include the apple (Malus domestica), whose centromeres show enrichment of the Hodor and Copia-7 LTR retroelements (Daccord et al. 2017; Zhang et al. 2019), and Bryco Copia elements in the centromeres of the moss Physcomitrella patens (Bi et al. 2024). Outside of land plants, the green algae Chlamydomonas reinhardtii has centromeres composed of ZeppL LINE retroelements (Craig et al. 2021, 2023). In animals, retrotransposons also show strong associations with centromeres, including LINE elements in humans (Logsdon et al. 2021; Hoyt et al. 2022), KERV endogenous retroviruses in kangaroo (Ferreri et al. 2011), and Jockey LINE elements in Drosophila (Chang et al. 2019; Courret et al. 2023). Hence, diverse families of centrophilic retroelements are observed across eukaryotic species, potentially representing multiple instances of convergent centrophilic adaptation.
In plant genomes, the predominant association between transposons and centromeres involves retrotransposons. However, in fungi and animals, a notable connection exists between Pogo DNA elements and the centromeres. The mammalian CENP-B protein and the fission yeast Abp1, Cbh1, and Cbh2 proteins are centromere-enriched and show sequence similarity to Pogo-type transposases (Kipling and Warburton 1997). Mammalian CENP-B binds as a homodimer to a 17-bp DNA sequence (CENP-B box) found within alpha-satellites of multiple species (Masumoto et al. 1989; Tanaka et al. 2001). Although Cenpb mutants do not have strong effects on endogenous centromere function (Hudson et al. 1998), de novo centromere formation on human artificial chromosomes requires CENP-B (Okada et al. 2007). It has further been speculated that CENP-B may retain the ability to generate DNA nicks or breaks and thereby promote centromere recombination (Kipling and Warburton 1997). However, to date, Pogo-derived centromeric proteins have yet to be reported in plants.
Holocentric and metapolycentric chromosome architectures
In many plants and animals, a derived state of holocentricity has evolved, in which chromosomes display multiple points of kinetochore attachment, distributed along the length of the chromosomes (Fig. 1F,G; Table 1; Melters et al. 2012; Heckmann et al. 2013; Drinnenberg et al. 2014; Marques et al. 2015; Hofstatter et al. 2022; Kuo et al. 2023; Neumann et al. 2023). The transition to holocentricity has evolved independently at least 13 times across the eukaryotic radiation, with at least four origins in angiosperms (Droseraceae, Convolvulaceae, Melanthiaceae, and Juncaceae/Cyperacea), and on multiple occasions within arthropods and nematodes (Melters et al. 2012). Attachment of microtubules along the length of the chromosome has the potential to facilitate genome evolution. For example, holocentric chromosome fragments and fusions may be more stable through cell division compared with acentric, dicentric, or polycentric chromosomes formed from rearrangement of monocentric genomes. Indeed, this is consistent with observed patterns of holocentric genome evolution, including frequent end-to-end chromosome fusions in Rhyncospora and the Lepidoptera (Hofstatter et al. 2022; Wright et al. 2024). Holocentricity also presents challenges during meiosis, in which a single round of replication is coupled to two rounds of chromosome segregation (Villeneuve and Hillers 2001; McAinsh and Marston 2022). In monocentrics, centromere behavior is altered during meiosis-I, such that sister chromatid centromeres are mono-orientated to the same cell pole, allowing segregation of homologs during the first division (Villeneuve and Hillers 2001; McAinsh and Marston 2022). Because of multiple spindle attachment points in holocentrics, it is challenging to ensure centromere mono-orientation and robust segregation of homologs at meiosis-I. This has been solved via achiasmatic meiosis, or inverted meiosis, in which sister chromatids separate at meiosis-I and homologs separate at meiosis-II (Cabral et al. 2014; Hofstatter et al. 2021). Despite these challenges, holocentric chromosome architecture has evolved on multiple occasions, indicating a potentially widespread benefit.
Recently, complete genomes have been assembled from several plant holocentric lineages, providing new insights into their sequence architecture (Fig. 1F,G; Table 1). The sedge (Cyperaceae) and rush (Juncaceae) monocot families contain numerous holocentric lineages, although monocentric species are also known (Melters et al. 2012; Hofstatter et al. 2021). Holocentric Rhynchospora species (Cyperaceae) are typified by multiple arrays of 172-bp Tyba satellites that are distributed along the chromosomes and are CENH3-occupied (Fig. 1F; Table 1; Marques et al. 2015; Hofstatter et al. 2022). Tyba arrays are ∼15–25 kb in length and are spaced between ∼300 and 400 kb along the chromosomes in three Rhynchospora species (Hofstatter et al. 2022). CENH3 enrichment was observed to rise toward the center of each Tyba array, resembling the distributions observed in larger monocentric satellite arrays, such as in A. thaliana (Hofstatter et al. 2022; Wlodzimierz et al. 2023b). The Tyba arrays are invaded by CRRh chromovirus retrotransposons, showing the centrophilic invasion also occurs in repeat-based holocentromeres (Hofstatter et al. 2022). Interestingly, TCR1 and TCR2 nonautonomous helitrons are also associated with Tyba repeats and have been implicated in spread of the satellite arrays along the chromosomes (Hofstatter et al. 2022). Animals are also known with repeat-based holocentric organization, for example, in the nematode genus Meloidogyne (Despot-Slade et al. 2021).
Chionographis japonica is a monocot belonging to Melanthiaceae, which has holocentric chromosomes, each with seven to 11 evenly spaced megabase-scale satellite arrays (average, 1.89 Mb) of Chio1 and Chio2 monomers that are CENH3-occupied (Fig. 1G; Table 1; Kuo et al. 2023). Hence, the C. japonica repeat arrays are larger than in Rhynchospora, although the Chio monomer repeats are shorter (23 and 28 bp) than Tyba (Fig. 1G; Table 1; Hofstatter et al. 2022; Kuo et al. 2023). In contrast to Rhynchospora and Chionographis, the rush Luzula elegans (Juncaceae), while possessing holocentric chromosomes with multiple kinetochore attachment points, does not show a clear association between centromere position and satellite or retrotransposon repeats (Heckmann et al. 2013, 2014; Nagaki et al. 2005a). In this case, epigenetic marks may specify centromere location. For example, in the holocentric nematode C. elegans, CENP-A loading associates with gene-associated chromatin states rather than repeated sequences (Gassmann et al. 2012; Steiner and Henikoff 2014). Further diversity in plant holocentric architecture was revealed in Cuscuta species, which have lost the kinetochore gene KNL2 and in which CENH3 is enriched in heterochromatin, and yet, microtubules attach along the entire length of the chromosome (Vondrak et al. 2021; Oliveira et al. 2022; Neumann et al. 2023). Similarly, loss of CENP-A underpins evolution of holocentricity in four insect lineages (Hemiptera, Ephemeroptera, Odonata, and Lepidoptera) (Drinnenberg et al. 2014). These patterns are consistent with recurrent evolution of holocentric chromosomes in independent lineages via different molecular mechanisms and involving distinct DNA sequences and chromatin states.
A further class of centromere architecture has been observed in leguminous plants, for example, in Pisum and Lathyrus species, which are distinguished cytologically by extended primary constrictions, comprising up to a third of metaphase chromosomes and possessing multiple distinct domains of CENH3 chromatin (Neumann et al. 2015; Schubert et al. 2020; Macas et al. 2023). For example, recent work has assembled the 81.8-Mb centromeric region of Chromosome 6 of Pisum sativum (Fig 1H; Table 1; Macas et al. 2023). This region contains multiple megabase-scale satellite repeat families, only a subset of which are CENH3-occupied, whereas other sequence classes, including genes and transposons are evenly distributed across the region (Macas et al. 2023). Despite the CENH3-occupied satellites being separated by megabases of intervening sequences, immunocytology revealed a single cellular focus of CENH3, potentially indicating looping or clustering of the occupied arrays in the nucleus (Macas et al. 2023). Metapolycentric may represent evolutionary transition states between monocentric and holocentric architectures. Indeed, high levels of satellite diversity have been documented in the plant tribe Fabeae, which includes the genera Pisum, Lathyrus, Vicia, and Lens, consistent with rapid evolutionary change of their centromeres (Ávila Robledillo et al. 2020).
DNA methylation and centromeric epigenetic states
Studies across eukaryotes have revealed that satellite-type centromeres are often DNA methylated, including in plants and humans (Vongs et al. 1993; Naish et al. 2021; Altemose et al. 2022; Gershman et al. 2022; Hofstatter et al. 2022; Hoyt et al. 2022; Macas et al. 2023; Wlodzimierz et al. 2023b). For example, the A. thaliana satellite repeats are densely DNA methylated in the CG context (Vongs et al. 1993; Naish et al. 2021; Wlodzimierz et al. 2023b). However, A. thaliana CEN178 satellites with high CENH3 occupancy show depletion of CHG context DNA methylation (Naish et al. 2021; Wlodzimierz et al. 2023b), which was also evident in soybean centromeres (Wang et al. 2023a). CHG DNA methylation in plants is maintained in an epigenetic loop with H3K9me2, such that in A. thaliana mutation of the KRYPTONITE/SUVH4 SET domain protein disrupts maintenance of both H3K9me2 and CHG DNA methylation (Jackson et al. 2002; Du et al. 2014; Stroud et al. 2014). As CENH3 cannot sustain H3K9me2, this provides an explanation for depletion of CHG DNA methylation coincident with the loading of CENH3 (Fig. 2C). This pattern is consistent with methylation within the Tyba arrays on holocentric Rhyncospora pubera chromosomes, which showed high CG methylation but were relatively depleted for CHG context DNA methylation (Hofstatter et al. 2022), which was also true for Chio repeats in C. japonica (Kuo et al. 2023). In contrast, in the metapolycentric Psium centromere 6, some CENH3-occupied tandem repeat arrays showed reduced CHG methylation, whereas others did not (Macas et al. 2023), and the retrotransposon-based centromeres of Triticum monococcum show depletion of CG-context methylation coincident with CENH3 loading compared with the rest of the chromosome (Ahmed et al. 2023). This is reminiscent of human CENP-A-enriched regions of the alpha-satellite arrays, which are CG-hypomethylated, in contrast to the surrounding unoccupied satellites that are densely DNA methylated (Miga et al. 2020; Logsdon et al. 2021, 2023; Altemose et al. 2022; Gershman et al. 2022). Hence, although centromeres are frequently heavily DNA methylated, the specific contexts and the relation to the CENH3/CENP-A-occupied sequences vary between species.
Recently, a functional link between DNA methylation and centromere identity has been made in Arabidopsis. VIM1 (an ortholog of mammalian UHRF1) encodes a conserved E3 ligase required for maintenance of CG DNA methylation, which is targeted to the Arabidopsis chromocenters (Bostick et al. 2007; Woo et al. 2007; Kraft et al. 2008). Mutants in VIM1 show hypomethylated CEN178 repeats, and cytological analysis showed that the satellite arrays are decondensed, with reduced CENH3 immunostaining signal (Bostick et al. 2007; Woo et al. 2007; Kraft et al. 2008). Recent work investigating CENH3-mediated genome elimination in A. thaliana has shown that vim1 mutations increase the frequency of haploid induction by CENH3-tailswap-GFP lines (Marimuthu et al. 2021). This implies that epigenetic change caused by vim1 increases the functional mismatch between centromeres, leading to greater chromosome missegregation and higher rates of genome elimination (Marimuthu et al. 2021). Together, this suggests a potential role for VIM1 and/or CG methylation in the recruitment or stabilization of CENH3 on the satellite arrays, potentially by impacting the ability of the inner kinetochore proteins to oligomerize and compact the centromeric chromatin (Fig. 2C,D).
In plants, DNA methylation is frequently used as a mechanism to limit transposable element mobility and can be guided by short interfering RNAs (siRNAs) via the RNA-directed DNA methylation (RdDM) pathway (Law and Jacobsen 2010; Gutbrod and Martienssen 2020). Consistent with this, A. thaliana centrophilic ATHILA elements are densely methylated in all sequence contexts and accumulate high levels of 21–24 siRNAs (Creasey et al. 2014; Lee et al. 2020; Naish et al. 2021; Wlodzimierz et al. 2023b). Interestingly, in T. monococcum, although the body of centromeric Cereba and Quinta elements are highly DNA methylated, their LTRs were hypomethylated (Ahmed et al. 2023). As the LTRs contain promoters for these types of transposons, this is consistent with ongoing Cereba and Quinta centrophilic transposition in wheat. Cereba and Quinta retroelements are also CENH3-enriched, in contrast to A. thaliana ATHILA centromeric insertions, which are CENH3-depleted relative to the surrounding satellites (Naish et al. 2021; Ahmed et al. 2023; Wlodzimierz et al. 2023b). Currently, siRNAs homologous to centromere satellites have not been widely reported in plants, although signal has been detected for the A. thaliana CEN178 repeats using small RNA northern blots (May et al. 2005). Strand-specific transcripts from the Arabidopsis satellites were also detected (May et al. 2005), which could be generated by RNA polymerase II, or the heterochromatic polymerases RNA Pol IV and V (Wendte and Pikaard 2017). Genome-wide mapping of R-loops, which are stable DNA:RNA hybrid structures, has also identified their signal in the maize centromeres (Liu et al. 2021), which points toward transcriptional activity. Indeed, analysis of RNA associated with maize CENH3 following ChIP showed enrichment of CRM retrotransposons and the CentC satellite repeats (Topp et al. 2004). Currently, the role of transcription in plant centromeres, the production of small RNAs, and how they interact with chromatin states and CENH3 loading are incompletely understood. Interestingly, simultaneous loss of DDM1 and RDR6 in the Col-0 accession of A. thaliana causes aneuploidy of Chromosome 5, which has a high density of ATHILA5 within its centromere (Shimada et al. 2023). The simultaneous loss of ddm1 and rdr6 causes a strong up-regulation of retrotransposon expression (Lee et al. 2020), indicating that heavily ATHILA-invaded centromeres are dependent on siRNA-mediated epigenetic silencing for normal function during cell division.
Together with dense DNA methylation, centromeres typically share other features of classical heterochromatin, including cytological condensation and late DNA replication (Schubert et al. 2020; Wear et al. 2020). It should also be noted that many eukaryotes, including budding yeast, fission yeast, D. melanogaster, and C. elegans lack detectable DNA cytosine methylation, although other chromatin marks, including H3K9me3, can function to suppress transposons in its place (Gutbrod and Martienssen 2020). Despite heterochromatin being a common feature of plant centromeric regions, there is currently limited consensus on how these epigenetic environments regulate centromere identity and function. For example, analysis of DNA methylation mutants in A. thaliana has not revealed strong defects on chromosome segregation (Yelina et al. 2012). This is in contrast to work in fission yeast, in which RNAi guides heterochromatin (H3K9me3) to DNA repeats that flank the core centromere and which is required for normal centromere function and cohesion (Hall et al. 2003; Volpe et al. 2003). Further work will be required to understand the wider roles of heterochromatic marks and small RNAs in plant centromeres across the different architectural types.
What drives rapid centromere evolution?
As noted, centromeres are characterized by rapid DNA sequence and kinetochore protein evolution within and between species (Malik and Henikoff 2009; Rhind et al. 2011; Melters et al. 2013; Logsdon et al. 2023; Wlodzimierz et al. 2023b), which presents a paradox given their conserved cellular function (Henikoff et al. 2001). An influential model to explain rapid centromere evolution is the drive hypothesis (Malik 2009; Akera et al. 2019; Finseth 2023). This model considers female meiosis, as it occurs in animals and plants, in which only one of the four meiotic products survives to differentiate into an egg or gametophyte (Fig. 3A; Malik 2009; Akera et al. 2019). Selfish centromere sequences that are able to bias their inheritance into the surviving female gamete/spore have the potential to achieve a non-Mendelian transmission advantage to the next generation, creating an evolutionary arms-race (Fig. 3A; Malik 2009; Akera et al. 2019). Consistent with this theory, selfishly driving centromeres that act via female meiosis have been observed in mice and monkeyflowers (Fishman and Saunders 2008; Chmátal et al. 2014; Akera et al. 2017, 2019; Iwata-Otsubo et al. 2017; Kumon et al. 2021).
In the case of monkeyflower, interspecies hybrids between Mimulus guttatus and Mimulus nastutus show highly distorted inheritance (>80%) of the D centromere haplotype on Chromosome 11 (Fishman and Willis 2005; Fishman and Saunders 2008). The molecular basis of D drive is not known but correlates with the driving chromosome having larger cytological Cent728 satellite repeat FISH signal (Fishman and Saunders 2008). In this scenario, a larger satellite array may recruit more CENH3 or load it in a “stronger” arrangement that leads to biased segregation into the functional female megaspore. In mice, natural variation in satellite repeat arrays that cause drive is linked to differential histone phosphorylation and recruitment of microtubule destabilizing factors (Iwata-Otsubo et al. 2017; Akera et al. 2019; Kumon et al. 2021). As a consequence, the driving mouse centromeres detach from spindle microtubules more frequently during meiosis and oogenesis, which reduces their segregation into the polar bodies (Iwata-Otsubo et al. 2017; Akera et al. 2019; Kumon et al. 2021). Therefore, variant centromere DNA sequences that can bias their inheritance during female meiosis have the potential to outcompete other sequences. This potentially causes an evolutionary arms-race whereby competition between selfish centromere sequences drives rapid evolution.
It has further been proposed that centromere arms-races would select for neutralizing or balancing mutations in trans-acting factors, including CENP-A/CENH3, to mitigate the effects of selfish DNA variants, which is consistent with rapid amino-acid divergence of these proteins observed between species (Malik 2009; van Hooff et al. 2017; Kursel and Malik 2018). However, in A. thaliana very few segregating CENH3 polymorphisms were observed in a sample of 66 diverse accessions, despite extensive structural variation in the CEN178 satellite arrays (Wlodzimierz et al. 2023b). The pace of centromere DNA coevolution may be further accelerated relative to the chromosome arms by elevated mutation rates and specific modes of homogenizing recombination that could be coupled to the presence of CENH3 and the kinetochore (Fig. 3B,C; Dover 1982; Rudd et al. 2006; Miga and Alexandrov 2021; Logsdon et al. 2023; Wlodzimierz et al. 2023b). It remains unclear which aspects of plant female meiosis are polar and capable of providing information for selfish centromeres to exploit. As female meiosis occurs in plant ovules, developmental or hormonal gradients within these organs may be instrumental in setting up cellular and/or spindle polarity that can be targeted during drive by selfish centromeres (Fig. 3A).
Maize abnormal Chromosome 10 (Ab10) is a further instructive example of meiotic drive (Birchler et al. 2003; Dawe 2022). Ab10 is a variant of the endogenous Chromosome 10 that bears large megabase-scale tandem repeat arrays, which are cytologically evident as heterochromatic “knobs” (Dawe et al. 2018; Swentowsky et al. 2020). In heterozygotes, Ab10 shows strong driving inheritance (∼83%) through female meiosis (Higgins et al. 2018). Interestingly, the knob does not recruit CENH3 or the kinetochore, and instead, drive requires an array of Kinesin driver (Kindr) genes, which act on the knob180 tandem repeats, and TR-1 Kinesin, which acts on the TR-1 359-bp tandem repeats (Dawe et al. 2018; Swentowsky et al. 2020). These kinesin/tandem repeat drive systems generally act synergistically but can also act independently (Dawe et al. 2018; Swentowsky et al. 2020). This indicates that the Ab10 system is circumventing the canonical centromere machinery, which also has implications for holocentric evolution in which species have lost CENH3 or kinetochore proteins (Drinnenberg et al. 2014; Neumann et al. 2023). Importantly, Ab10 knobs can have negative effects on male and female fitness when homozygous (Higgins et al. 2018). Similarly, female drive of D haplotypes in the monkeyflower is associated with a decrease in male reproductive fitness (pollen viability) (Fishman and Saunders 2008; Fishman 2013), indicating fitness trade-offs that may prevent fixation of the driving centromeres. It is also possible that centromere haplotypes could display conditional drive, such that variant A beats B, B beats C, and C beats A, which has the potential to maintain sequence diversity. Although drive during female meiosis is a compelling model to explain rapid centromere evolution via competitive arms-races, other factors should be considered. For example, centromere evolution is also rapid in isogamous species, such as fission yeast (Rhind et al. 2011), indicating other mechanisms likely exist. In this respect, it is relevant to consider haploid-induction by CENH3 variants in plants, which also provides a potential mechanism for rapid chromosome loss and genome restructuring during evolution (Ravi and Chan 2010; Karimi-Ashtiyani et al. 2015; Kuppu et al. 2015; Tan et al. 2015; Lv et al. 2020; Wang et al. 2021).
As diverse retrotransposons display centrophilic adaptation, it is possible that these sequences are engaged in drive themselves or that they modify drive. As centromeric recombination in satellite arrays can act to purge retrotransposons (Shi et al. 2010; Wlodzimierz et al. 2023b), this may represent a defense against drive (Fig. 3C). Indeed, theoretical work supports that recombination during meiosis may have evolved as a “scrambling defense” against driving elements that link themselves to cis-acting centromere sequences (or become them) (Haig and Grafen 1991). In this scenario, the instability of centromeres could represent an adaptation to prevent establishment of driving sequences. Holocentricity could also be considered as a defense against drive, as multiple centromere locations will potentially be more robust to manipulation by driving agents. Because of species showing varying centromere composition by satellites versus retrotransposons, it seems likely that mechanisms exist in which genomes can evolve between these states (Fig. 3C). It is easy to conceive how a successful centrophilic retrotransposon could colonize and take over a satellite-based centromere, such as observed in Einkorn wheat and Drosophila (Ahmed et al. 2023; Courret et al. 2023). Yet, how centromere satellite arrays can emerge from a transposon-based architecture remains unclear, although evidence exists for formation of tandem repeats from centrophilic retrotransposons in plants (Cheng and Murata 2003; Ito et al. 2004; Sharma et al. 2013), CR1-C retrotransposons in chicken (Shang et al. 2010), and centromeric KERVs in kangaroo (Koga et al. 2023). The pathways that form such transposon-derived tandem repeat arrays are poorly characterized but provide a potential route for satellite arrays to evolve from transposon-dominated centromeres.
Centromeres have a potent suppressive effect on meiotic crossover within themselves and the surrounding sequences (Mahtani and Willard 1998; Vincenten et al. 2015; Nambiar and Smith 2018; Langley et al. 2019; Naish et al. 2021; Fernandes et al. 2024), which can shape genetic variation and genome evolution. For example, monocentric plant chromosomes show pronounced telomere–centromere gradients of gene density, transposon diversity, and epigenetic modifications (Rowan et al. 2019; Naish et al. 2021; Ahmed et al. 2023; Chen et al. 2023; Fernandes et al. 2024). As similar gradients are not evident in holocentric Rhynchospora genomes (Hofstatter et al. 2022), this argues for a dominant role in monocentric architecture in influencing multiple other aspects of genome organization. However, despite showing a uniform landscape of gene, repeat, and epigenomic patterns, Rhynchospora shows distalized meiotic crossover frequency, potentially indicating a role for the telomere bouquet (Castellani et al. 2024). As a consequence of centromere-proximal recombination–suppression in monocentric species, the genes and transposons located there will tend to maintain linkage and coinherit, with the potential to form supergenes (Finseth et al. 2022). For example, the driving D centromere haplotype in Mimulus spans ∼12 Mb, contains at least 350 genes, and occurs at varying frequency in natural populations (Fishman 2013; Finseth et al. 2022). These regions could also contain trans-acting modifiers of centromere inheritance, such as the Kindr repeats in maize (Dawe et al. 2018; Swentowsky et al. 2020) and ATHILA elements in Arabidopsis (Swentowsky et al. 2020; Shimada et al. 2023). As further centromere regions are fully assembled, it will be interesting to understand the extent to which linked-sequences contribute to centromere identity, drive, and other genomic processes.
Prospects for engineering synthetic centromeres
Plant centromeres are not defined by conserved DNA sequences, either within genomes, within species, or across species. This sequence lability, as well as the role of epigenetic factors, makes engineering centromeres inherently difficult. However, as new genome assemblies accumulate with complete centromeres, a deeper understanding will be achieved of the sequences and chromatin states that robustly confer centromere identity to plant chromosomes. Experiments to re-engineer centromeres, including the application of CRISPR-Cas9, will be powerful to define neocentromeres and minimal repeat arrays capable of maintaining chromosomes in vivo. For example, CRISPR guide RNAs targeting Cas9 to chromosome-specific alpha-satellite repeats are effective at creating aneuploidy in human cells (Bosco et al. 2023), whereas CRISPR targeting at sites flanking human centromere 4 created an acentric chromosome and triggered neocentromere formation (Murillo-Pineda et al. 2021). Therefore, similar CRISPR strategies in plants could be used to generate reduced, expanded, or rearranged centromeres; acentrics; neocentromeres; or other chromosome rearrangements and novel karyotypes. Similarly, work targeting CENH3 via LexA fusions in maize shows that this strategy can also lead to dicentrics, chromosome instability, and formation of autonomously replicating mini-chromosomes (Dawe et al. 2023). More precise knowledge of what controls centromere identity in plants, at the genetic and epigenetic levels, may provide tractable sequences that can be synthesized in artificial chromosomes. Ideally, relatively small sequence arrays that confer strong centromere identity are required to avoid the need to synthesize multimegabase satellite arrays. For example, the Rhynchospora Tyba arrays provide examples of ∼15- to 25-kb blocks of satellites that are capable of CENH3 recruitment and kinetochore assembly (Hofstatter et al. 2022). Ultimately, synthetic chromosomes have the potential to act as a vector to deliver complex, multigene pathways into plant species. These advanced forms of genetic modification have the potential to provide powerful new tools for adapting and engineering crops to the changing climate.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank Jiří Macas for discussions and James Higgins, Teri Mandáková, and Martin Lysak for generating the cytogenetic data shown in Figure 1, B and C (Lambing et al. 2020; Naish et al. 2021). Figure 3, B and C, is reproduced from Wlodzimierz et al. (2023b). This work was supported by Biotechnology and Biological Sciences Research Council grants BB/S006842/1, BB/S020012/1, and BB/V003984/1; European Research Council Consolidator Award ERC-2015-CoG-681987; and Human Frontier Science Program award RGP0025/2021 to I.R.H., and a Broodbank Fellowship to M.N.
Footnotes
-
Article published online before print. Article and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278409.123.
-
Freely available online through the Genome Research Open Access option.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.
References
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵
- ↵