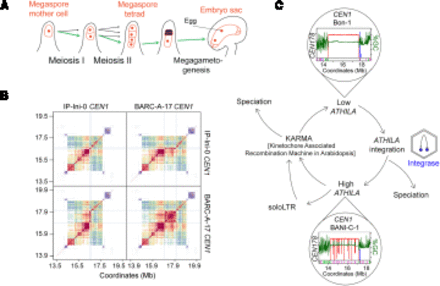
Centromere drive and evolution in plants. (A) Diagram showing differentiation of the megaspore mother cell (MMC) in a subapical position of an Arabidopsis ovule (red). The MMC undergoes meiosis resulting in four haploid daughter cells, three of which enter programmed cell death, leaving a functional megaspore that differentiates into the haploid embryo sac. The embryo sac contains the egg cell, which is fertilized by pollen to produce progeny seed. The green arrows indicate the theoretical path a selfish centromere could follow to overtransmit itself in a non-Mendelian manner, causing meiotic drive. (B) StainedGlass sequence identity heat maps comparing CEN1 within and between IP-Ini-0 (Spanish) and BARC-A-17 (French) accessions of A. thaliana (Vollger et al. 2022; Wlodzimierz et al. 2023b). Red and orange indicate high levels of sequence similarity. The gray lines demarcate a ∼1-Mb BARC-A-17 region that is similar to a ∼100-kb IP-Ini-0 region, indicating centromere sequence dynamics since these accessions diverged (Wlodzimierz et al. 2023b). (C) A cyclical model for centromere evolution in A. thaliana, which we propose alternates between high and low ATHILA retrotransposon invasion states, represented by CEN1 in the Bon-1 and BANI-C-1 accessions respectively, which belong to the same satellite similarity group, yet differ in the level of ATHILA invasion (Wlodzimierz et al. 2023b). ATHILA transcription generates new copies with the potential to integrate into the centromeres. Intact ATHILA may undergo internal recombination to generate soloLTRs. Simultaneously, satellite arrays undergo recombination, resulting in repeat homogenization and purging of ATHILA via a proposed kinetochore associated recombination machine in Arabidopsis (KARMA) (Miga and Alexandrov 2021; Wlodzimierz et al. 2023b). Inter-species satellite and retrotransposon polymorphisms are also consistent with roles of centromere evolution in speciation.











