A single-cell transcriptome atlas of the maturing zebrafish telencephalon
Abstract
The zebrafish telencephalon is composed of highly specialized subregions that regulate complex behaviors such as learning, memory, and social interactions. The transcriptional signatures of the neuronal cell types in the telencephalon and the timeline of their emergence from larva to adult remain largely undescribed. Using an integrated analysis of single-cell transcriptomes of approximately 64,000 cells obtained from 6-day-postfertilization (dpf), 15-dpf, and adult telencephalon, we delineated nine main neuronal cell types in the pallium and eight in the subpallium and nominated novel marker genes. Comparing zebrafish and mouse neuronal cell types revealed both conserved and absent types and marker genes. Mapping of cell types onto a spatial larval reference atlas created a resource for anatomical and functional studies. Using this multiage approach, we discovered that although most neuronal subtypes are established early in the 6-dpf fish, some emerge or expand in number later in development. Analyzing the samples from each age separately revealed further complexity in the data, including several cell types that expand substantially in the adult forebrain and do not form clusters at the larval stages. Together, our work provides a comprehensive transcriptional analysis of the cell types in the zebrafish telencephalon and a resource for dissecting its development and function.
The telencephalon has important functions in learning, memory, social behavior, and decision making (Aoki et al. 2013; Cheng et al. 2014; Lal et al. 2018; Stednitz et al. 2018). Developing from the rostral-most part of the neural tube in zebrafish, the telencephalon consists of three major structures—the olfactory bulb, dorsally located pallium, and ventrally located subpallium (Fig. 1A). Unlike the mammalian telencephalon, the zebrafish telencephalon has an everted morphology that consists of two lobes separated by a T-shaped ventricle (Folgueira et al. 2012). Previous anatomical studies have associated regions in the everted zebrafish telencephalon with those in the evaginated mammalian forebrain (Supplemental Fig. S1A; Porter and Mueller 2020). In the zebrafish, subdivisions of the pallium and subpallium have been proposed based on gene expression analysis of some key markers such as emx1, emx2, emx3, eomesa, tbr1b, and others in the pallium (Ganz et al. 2014) and dlx2a, dlx5a, and others in the subpallium (Ganz et al. 2012). However, it has been unclear how many transcriptionally defined neuronal cell types are represented in these subdivisions. Previous work on brain-wide single-cell RNA sequencing (RNA-seq) of the juvenile brain highlighted approximately four transcriptionally distinct clusters in the pallium (Raj et al. 2018). However, focused regional analysis can uncover additional heterogeneity in brain areas, as was observed in our published work on the zebrafish habenula (Pandey et al. 2018). Such detailed single-cell RNA-seq data sets can support cross-species comparisons (Hashikawa et al. 2020) and functional studies with discovered cell types (Stednitz et al. 2018; Ncube et al. 2022). Therefore, a more detailed survey of the neuronal cell types in the pallium and subpallium is likely to reveal previously unrecognized cell types and provide a resource for future studies.
Integrated graph clustering of telencephalic cells. (A) Schematic for the stages profiled and the scRNA-seq using the 10x Genomics platform. (B) UMAP representation of the integrated analysis across 6-dpf, 15-dpf, and adult zebrafish telencephalon, faceted and colored by age. (C) UMAP representation of the integrated analysis of the zebrafish telencephalon, colored by annotated and interpreted cell types. The right panel displays gene expression profiles of select broad marker genes for identification of neuronal (pallium and subpallium), nonneuronal, and progenitor subtypes within the data set. (IN) Immature neuron.
The telencephalon undergoes growth, neurogenesis, and functional maturation from the larval stages of development to adulthood. In parallel, several behaviors mediated by neuronal ensembles in the pallium and subpallium emerge, including social behaviors, memory, and fear learning (Aoki et al. 2013; Cheng et al. 2014; Lal et al. 2018; Stednitz et al. 2018). It is conceivable that neurons involved in these behavioral repertoires arise later in development. Recent work dissected the emergence of neuronal cell types in the entire brain, providing a broad overview of the diversification of neurons and progenitors until the late larval stages (Raj et al. 2020). However, a systematic, detailed, and focused comparison of the larval and adult telencephalic cell types is still missing.
The aim of this study is to use single-cell RNA-seq across life stages to obtain a comprehensive picture of the transcriptionally distinct cell types of the zebrafish pallium and subpallium.
Results
Integrated graph clustering identifies cell types in the zebrafish telencephalon
To analyze the molecular features of the cells from the zebrafish telencephalon, we isolated and performed single-cell RNA-seq on the telencephalons of 6-day-postfertilization (dpf), 15-dpf, and adult zebrafish in nine, four, and two experimental batches, respectively (Fig. 1A; Supplemental Figs. S1B, S2). At 6 and 15 dpf, we dissected the telencephalon, including the pallium, subpallium, and olfactory bulb, as well as regions of the preoptic area that were continuous with the subpallium. In the adult, the olfactory bulb was removed during dissection to retain more single-cell coverage in the two main regions of interest: the pallium and subpallium. We collected a total of approximately 64,000 cells across these three different ages using the 10x Genomics platform. Quality filtering resulted in a total of 21,247 cells from the 6-dpf, 15,476 cells from 15-dpf, and 27,125 cells from the adult telencephalon (Fig. 1B).
To integrate the cells from the three different time points and identify shared and divergent clusters, we used Seurat's data integration pipeline, which combines canonical correlation analysis (CCA) and mutual nearest neighbor (MNN) analysis (Stuart et al. 2019). After integration, the cells were clustered and labeled based on the marker genes conserved across all three time points. Of the 38 clusters identified in the integrated analysis of the telencephalon, 26 clusters were neuronal, and 12 clusters were nonneuronal (Fig. 1C).
Among the nonneuronal cells, we found clusters expressing prototypical markers for progenitors, microglial cells, oligodendrocytes, and endothelial cells (Fig. 1C, side panels; Supplemental Table S1). The progenitors included radial glial cells (Lange et al. 2020) that express markers such as fabp7a and mdka, cycling progenitors that express markers such as pcna, and proneural precursors that express markers such as neurog1 (Supplemental Fig. S3). Of two progenitor pools, Progenitor_01 is the larger and labeled by fabp7a, a gene known to be expressed in the telencephalic ventricular zone in adult zebrafish (Adolf et al. 2006). The smaller Progenitor_02 cluster is labeled by fabp7b, and the differential expression (DE) of these two genes was previously described (Liu et al. 2004). We also found a cluster that expressed canonical mammalian astrocyte markers such as slc1a3b and slc1a2b, highlighted by a recent zebrafish study (Chen et al. 2020) and identified as dormant radial glia by past studies (Lange et al. 2020). In addition, these putative astrocytes are enriched in mfge8a, a marker reported to be specific for telencephalic astrocytes in mice (Zeisel et al. 2015). Based on RNA-FISH of cspg5b, a gene also enriched in this cluster, we spatially localized the cells to the medial region of the telencephalon lining the ventricles (Supplemental Fig. S4).
To assign an approximate regional identity to the mature neuronal clusters, we identified a host of markers for each (Supplemental Table S1). A large majority of clusters were defined by at least one gene that was a largely specific marker of that cell type (Fig. 2). We assigned clusters as belonging to pallium and subpallium based on the expression of known markers of these regions (Mueller et al. 2008; Ganz et al. 2012, 2014). Consistent with expression patterns in mammals, dlx1a, dlx2a, dlx2b, dlx5a, dlx6a, tbr1b, and neurod1 distinguish between the zebrafish pallium and subpallium. Among the neuronal clusters, we also found one cluster belonging to the olfactory bulb (Supplemental Fig. S4), two clusters belonging to the habenula (Pandey et al. 2018), and two belonging to the preoptic area. These clusters, established based on our previously published work or their spatial location in situ, were not analyzed further.
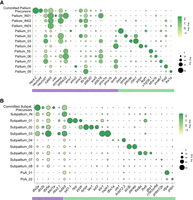
Marker genes of the neuronal cell types in the larval telencephalon. (A) Expression profiles of marker genes that are specific or enriched in the pallium subtypes. (B) Expression profiles of marker genes that are specific or enriched in the subpallium subtypes. The purple bar on the bottom represents previously defined markers that were used to subcategorize the region, and the green bar represents markers identified by the integrated analysis. For the expression of additional genes, see Supplemental Figure S5 and Supplemental Table S1.
We assigned nine neuronal clusters to the pallium, the dorsal region of the telencephalon that is thought to contain structures homologous to the mammalian cortex, hippocampus, and some sections of the amygdala (Cheng et al. 2014; Ganz et al. 2014). Previously, the adult zebrafish pallium was subdivided into subregions such as the dorsal, medial, ventral, and lateral based on the combinatorial expression of marker genes such as eomesa, emx1, emx2, and emx3 (Ganz et al. 2014). Most of these genes were broadly expressed across multiple clusters in our data set (Fig. 2A). Other broad pallium markers observed in our data set included bhlhe22, rprml, and zbtb18.
These previously described broad markers were used to ascribe a proposed regional identity for some pallium clusters (Fig. 2; Supplemental Figs. S1A, S5). Cells in clusters Pallium_02 and Pallium_03 are both eomesa+ and express low levels of emx1. Pallium_02, unlike Pallium_03, is also characterized by its expression of emx3. These expression patterns are consistent with a region near the posterior subdivision of the dorsal pallium (Dp) or the adjacent ventral subdivision of the lateral zone of the dorsal pallium (Dlv) (Ganz et al. 2014). Clusters Pallium_05 and Pallium_06 are heterogeneous (Supplemental Fig. S5). The majority of cells in Pallium_05 and Pallium_06 represent at least two subtypes of glutamatergic cells marked by expression of either c1ql3b.1 or efna1b. The efna1b+ cell type expresses eomesa but not emx1, emx2, or emx3, whereas the c1ql3b.1+ cell type has low expression of eomesa, emx1, emx2, and emx3. These expression patterns indicate localization of these cell types to the dorsal subdivision of the lateral zone of the dorsal pallium (Dld) that may be related to the hippocampus (Ganz et al. 2014). The remaining cells in Pallium_05 are eomesa−/emx3+, which is consistent with expression in the medial zone of the dorsal pallium (Dm) (Ganz et al. 2014), the proposed pallial amygdala. It is unclear whether the excitatory neurons in Pallium_05 and Pallium_06 occupy discrete regions of the telencephalon or whether cells in these clusters represent cell types shared between regions.
Cells in the remaining pallial clusters were identified based on markers from a recent paper describing the teleostean amygdaloid complex (Porter and Mueller 2020). The strong expression of pvalb7+ in Pallium_07 is consistent with this cluster contributing to the zebrafish “integrative olfactory pallium” (IOP) and the medial pallium or dorsolateral pallial territory (Dl), which is a putative hippocampal homolog (Porter and Mueller 2020). The pyya+ Pallium_01 cluster may also contribute to the IOP. Pallium_01 has some pvalb7 expression and the strongest emx1 expression of all clusters; in adult zebrafish, emx1 is expressed only in Dp, consistent with these cells contributing to the IOP. Cells in clusters Pallium_04, Pallium_08, and Pallium_09 express members of the LIM homeobox family, including lhx9, lhx2b, lhx5, and lhx1a. The zebrafish genes lhx5 and lhx1, as well as tbr1b, mark the thalamic eminence (Wullimann and Mueller 2004; Turner et al. 2016), suggesting that cells in these clusters may derive from this structure. A subset of cells in Pallium_04 coexpresses lhx5 and otpa, which is consistent with the recently identified posterior division of the medial amygdala (Porter and Mueller 2020). The gene emx2 is also expressed in Pallium_08 and a subset of cells in Pallium_09, which is exclusively expressed in the central division of the pallium (Dc) in the adult telencephalon (Ganz et al. 2014).
In addition to the mature neuronal clusters, we also identified three immature neuron (IN) clusters in the pallium that were all enriched in markers such as tubb5 (Supplemental Fig. S4) but likely contribute to different mature neuron clusters. The committed pallium precursors are distinguished from these IN clusters by the coexpression of progenitor genes such as her4.2 (Fig. 2A). Pallium_IN01 was enriched in the expression of bhlhe22, a marker that is broadly expressed in Pallium_05, Pallium_06, and Pallium_07, suggesting that these INs contribute to these three pallium clusters. Pallium_IN02 is eomesa+, bhlhe22− and likely contributes to the eomesa+, bhlhe22− Pallium_02 and Pallium_03. Pallium_IN03 expresses tbr1b but not the pallium markers eomesa, bhlhe22, emx1, emx2, or emx3. This pattern of transcription factor expression most closely resembles the remaining transcripts in the mature clusters Pallium_08 and Pallium_09 (Fig. 2A). Pseudotime analysis supports the divergent developmental trajectories of Pallium_IN01 and Pallium_IN02, as these cells differentiate into Pallium_06 and Pallium_02 clusters, respectively (Supplemental Fig. S6). Defining gene modules that change as a function of pseudotime uncovered new marker genes dividing the telencephalic neuron types (Supplemental Fig. S7). In contrast to the multiple subtypes of INs in the pallium, a single IN cluster was present in the subpallium (Fig. 2B).
We identified eight mature neuronal clusters belonging to the subpallium, the ventral region of the telencephalon, which is thought to contain structures homologous to the mammalian basal ganglia (Mueller et al. 2008). Previously, the zebrafish subpallium was divided based on the expression of genes such as dlx2a, dlx5a, and lhx6 (Mueller et al. 2008; Ganz et al. 2012). These genes do not mark single clusters, whereas our clustering subdivides the subpallium into transcriptionally distinct neuron types based on multiple new marker genes such as pdrm12b (Fig. 2B).
The zebrafish subpallium was found to contain GABAergic interneurons, a cholinergic population, cells with similarity to medium spiny neurons, and an excitatory septal cluster. Subpallium_01 expresses nkx2.1, lhx6, sst1.1, and npy, suggesting that this cluster represents somatostatin+, neuropeptide Y+ interneurons. The Subpallium_03 cluster expresses markers of parvalbumin+ interneurons, including nkx2.1, sox6, etv1, nxph1, and pvalb6. Subpallium_02 and Subpallium_05 express markers of striatal medium spiny neurons, such as tac1 (substance P), sp8a, synpr, penkb (enkephalin), six3b, cxcl14, and foxp1b (Aguda et al. 2021). These clusters also express the neuropeptides pyya and pyyb. Subpallium_06 expresses nkx2.1, lhx6, and the cholinergic markers lhx8a, gbx1, and isl1. This cluster likely represents a recently identified population of neurons that synthesizes both acetylcholine and GABA and is involved in social behavior in zebrafish (Ncube et al. 2022). Unlike other subpallium clusters, Subpallium_04 expresses the glutamatergic marker slc17a6b (vglut2) and low levels of the GABAergic markers slc32a1 (vgat), slc6a1b (gat), gad1b (gad67b), and gad2 (gad65). This cluster is defined by septal markers, including isl1, zic1, zic2a, zic4, and zic5 (Supplemental Fig. S5; Woych et al. 2022), as well as several unique marker genes (Fig. 2B). The Subpallium_07 cluster is heterogeneous, with subsets of cells expressing septal markers (zic1, zic2a, zic4, and zic5) (Inoue et al. 2007; Zeisel et al. 2018), cholinergic markers (nkx2.1, lhx6, lhx8a, and gbx1) (Sandberg et al. 2016), interneuron markers (calb1, calb2b, pvalb6, and sst1.1) (Bugeon et al. 2022), and the dopamine receptor drd2b. An unidentified subset of cells coexpresses sp8a, rgs4, and c16h2orf66.
Taken together, these results define neuronal subtypes of the pallium and subpallium and provide a catalog of markers for their further study.
Spatial localization of neuronal cell types in the zebrafish telencephalon
To determine the spatial location of the markers for a subset of these single-cell clusters and support their subpallial or pallial identity, we collected fluorescent in situ hybridization (FISH) data for some marker genes (Fig. 3; Supplemental Table S2) and registered these image stacks into a shared coordinate space (Supplemental Video S1). Similar to previously published work (Pandey et al. 2018), we found that the neuronal cell types in the pallium and subpallium were largely regionalized. By observing the larger domains within the pallium using broad markers such as bhlhe22, rprml, and zbtb18, we conclude that there are two distinct regions of the pallium along the anterior–posterior axis that encompass neuronal cell types identified by our analysis (Fig. 3A). The posterior half, defined by the expression of rprml, consists of neuronal cell types such as pdyn+ Pallium_04 and lhx9+ Pallium_08 that are located dorsally (Fig. 3A). Whereas pdyn occupies the medial region of the dorsal posterior pallium, lhx9 is located in the lateral half of the dorsal posterior pallium. Other posterior neuronal cell types include pyya+ Pallium_01 and prkcda+ Pallium_03, both of which are localized laterally along different Z-planes of the ventral pallium. In the bhlhe22+ anterior pallium, in correspondence with the cluster abundances in the single-cell data, Pallium_06 occupied the largest expression domain (Fig. 3A). The INs also occupy regionalized niches within the pallium (Fig. 3A). Published in situ data for some marker genes in the adult telencephalon (Supplemental Fig. S8; Diotel et al. 2015) are consistent with the larval data, adult subdivisions suggested by previous work (Ganz et al. 2014), and pseudotime clustering. Markers of Pallium_02 and Pallium_03 are found in Dp/Dlv, whereas markers of Pallium_05 and Pallium_06 are found in Dld, Dc, and Dm.
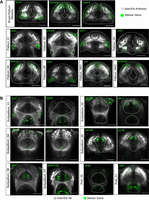
Spatial distribution of neuronal cell types in the larval telencephalon. (A) In situ expression patterns of cluster-specific marker genes for selected subclusters in the pallium at 10 dpf (dorsal view). RNA-FISH (green) was performed with a total-Erk (pale gray) costain for anatomy. Approximate demarcations of the pallium, based on the Z-Brain atlas masks (Randlett et al. 2015), are indicated by white dotted lines. (B) In situ expression patterns of cluster-specific marker genes for selected subclusters in the subpallium at 10 dpf (dorsal view). Approximate demarcations of the subpallium only (Subpallium_04, Subpallium_05, Subpallium_06) or subpallium and preoptic area (all other panels), based on the Z-Brain atlas masks (Randlett et al. 2015), are indicated by white dotted lines. The scale bar represents 50 µm. For the three-dimensional stack and depth information, see Supplemental Video S1.
Subpallium neuronal cell types were also regionalized along the anterior–posterior and dorso–ventral axis in the ventral part of the telencephalon. Neuronal clusters Subpallium_05 (penkb+) and Subpallium_02 (synpr+) were located in the anterior half of the subpallium along the dorso–ventral axis (Fig. 3B). In contrast, sst1+ Subpallium_01 is localized to the posterior half of the subpallial domain. Other neuronal clusters, including the prdm12b+ Subpallium_07, are localized to a band of neurons in the medial part of the subpallium along the anterior–posterior axis (Fig. 3B). We also localized the otpa+ and pdyn+ cluster of preoptic area neurons, PoA_01 and PoA_02, respectively, which were readily identified by their spatial position (Fig. 3B).
Together, these results provide validation and spatial localization for the majority of neuronal clusters found in the telencephalon through our single-cell analysis.
Comparison of neuronal cell types in the zebrafish and mouse forebrain
To further identify the zebrafish telencephalic neuron types, we compared zebrafish markers (Supplemental Fig. S5) to the mammalian literature (Supplemental Table S3) and performed computational integration with an adolescent mouse data set (Fig. 4A,B; Zeisel et al. 2018).
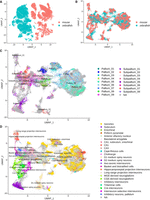
Integrated clustering of zebrafish and mouse forebrain cells. (A) UMAP representation of mouse and zebrafish mature neurons analyzed without integration. (B) UMAP representation of mouse and zebrafish mature neurons integrated with Harmony. (C) UMAP representation of the integrated analysis colored according to original identities in zebrafish samples. Gray cells (NA) represent mouse cells. (D) UMAP representation of the integrated analysis according to the original locations in mouse samples. Gray cells (NA) represent zebrafish cells.
Cells in clusters Pallium_02 and Pallium_03 were found to share expression of genes with the mouse subiculum (Supplemental Table S3), suggesting that Pallium_02 and Pallium_03 may represent the area between the hippocampus proper and the entorhinal cortex. The computational integration supports this designation (Fig. 4C,D), as these zebrafish cells cluster near mouse cells located in the subiculum and entorhinal cortex. Mouse orthologs of the markers of Pallium_02 and Pallium_03—Rprm (Pallium_02 only), Rspo2 (a subset of Pallium_03), and Cbln1 (both)—are expressed in the murine subiculum, piriform area, postpiriform transition area, piriform–amygdalar area, lateral amygdalar nucleus, basolateral amygdala, and entorhinal cortex (Supplemental Fig. S9; Supplemental Table S3). Together, expression of marker genes suggests that cells in clusters Pallium_02 and Pallium_03 share expression patterns with the murine subiculum, entorhinal cortex, and pallial amygdala.
Clusters Pallium_05, Pallium_06, and Pallium_07 are scattered among mouse cells located in the entorhinal cortex, isocortex, CA1, CA3, and piriform cortex (Fig. 4C,D). Pallium_07 is marked by high expression of pvalb7, which is expressed in the IOP region that has been proposed as the putative homolog of the mammalian entorhinal cortex (Porter and Mueller 2020) and the medial pallium (hippocampus homolog). Mouse orthologs of genes marking the efna1b+ population in the Pallium_05 and Pallium_06 clusters are expressed in the murine isocortex, hippocampus, and basolateral amygdala (Supplemental Figs. S5, S9). Cells expressing c1ql3b.1, another cell type found in Pallium_05 and Pallium_06, additionally express nptx1l, which is consistent with a report that NPTX1 coimmunoprecipitates with C1QL3 and that C1ql3 and Nptx1 are coexpressed in mouse cortical neurons (Sticco et al. 2021). This cell type is found in multiple mouse brain regions.
The emx2 expression in Pallium_08 and Pallium_09, marking these clusters at the Dc region, suggests that they may be located in a functional correlate of the mammalian cortex (Cecchi 2002; Ganz et al. 2014). However, we did not identify a population of cells that coexpresses emx2 and eomesa, which is also expressed in Dc (Supplemental Fig. S5). One hypothesis for these observations is that emx2+/eomesa− cells migrate to Dc from the thalamic eminence, whereas emx2−/eomesa+ cells derive from the dorsal proliferative matrix of the dorsal pallial division (Mueller et al. 2011). This hypothesis is strengthened by the expression of markers of Cajal–Retzius cells, which in mammals migrate from regions including the dorsal septum and thalamic eminence (Jiménez and Moreno 2022). The signature of the thalamic eminence, coexpression of Lhx1 and Lhx5, is also conserved between the mouse (Adutwum-Ofosu et al. 2016) and zebrafish. Adding further support to this classification, the computational integration clustered Pallium_08 near the murine isocortex and Cajal–Retzius cells in the UMAP plots (Fig. 4C,D), marked by expression of Emx2 (Supplemental Fig. S9). Determining whether emx2+ cells in Dc derive from the thalamic eminence will require further investigation.
Integration of the single-cell data sets between the two species suggested that the subpallium clusters were largely consistent between zebrafish and mouse. Cells in Subpallium_01 cluster near hippocamposeptal and long-range projection interneurons (Fig. 4C,D), which are Sst+ (Supplemental Fig. S9). In mice, the medial ganglionic eminence (MGE) gives rise to interneurons that express somatostatin or parvalbumin downstream from the transcription factors Nkx2.1 and Lhx6, also expressed in Subpallium_01 (Fig. 2B). Cells in Subpallium_03 cluster near MGE-derived neurogliaform and axo-axonic interneurons (Fig. 4C,D), which express Nxph1 (Supplemental Fig. S9). Interneurons derived from the caudal ganglionic eminence (CGE) in mammals, marked by coexpression of orthologs of Nr2f2 (Coup-TFII), Prox1, Sp8, Reln (reelin), Calb2 (calretinin), or Vip (vasoactive intestinal polypeptide), were not observed in the zebrafish telencephalon (Supplemental Fig. S5). Subpallium_06 clusters near cholinergic neurons that express Lhx8 (Fig. 4C,D), a transcription factor required for forebrain cholinergic neuron specification in mice (Zhao et al. 2003). As expected from their expression of striatum-like markers (Gokce et al. 2016) such as penkb+ and synpr+, Subpallium_02 and Subpallium_05 cluster near lateral ganglionic eminence (LGE)–derived striatal medium spiny neurons. Cells in Subpallium_02 cluster near D1 medium spiny neurons, which are marked by the expression of Tac1 (Supplemental Fig. S9). Subpallium_05 clusters near D2 medium spiny neurons and Cck interneurons and is marked by the expression of Penk. Orthologs of the dopamine receptors are minimally expressed in our data, with drd2b expressed substantially only in Subpallium_08. In addition to their presence in the zebrafish striatum equivalent, cells in Subpallium_02, Subpallium_05, and Subpallium_08 express markers indicating a potential contribution of these clusters to the LGE-derived nuclei of the extended amygdala (Porter and Mueller 2020). Cells in Subpallium_07 and Subpallium_08 are spread among multiple mouse cell types, including pallidal inhibitory neurons, basket and bistratified cells, cholinergic neurons, and interneuron-selective interneurons. Together, these analyses suggest that the cell types in the zebrafish subpallium generally resemble those in the mouse, whereas the cell types in the pallium are more divergent but can be mapped to plausible brain regions.
Comparison of the larval telencephalon to adult telencephalon in the integrated data
To assess the conservation and emergence of neuronal cell types from the larval to adult telencephalon, we investigated the changes in cellular proportions for the clusters identified by the integrated analysis across all three stages. Of the nine neuronal cell types we identified in the pallium, two expand in proportion from each representing ∼10% of the total pallium cells in larvae to >30% of the pallium in the adult telencephalon (Fig. 5A). These include the c1ql3b+ Pallium_05 and efna1b+ Pallium_06. Similarly, Pallium_07, which comprises only ∼3% of the total pallium cells in the 6-dpf animal, expands to ∼10% of the cells in the adult pallium. Pallium_07 represents a cluster of slc17a6a and slc17a6b+ excitatory neurons that are also marked by the expression of pvalb7 (Fig. 5B; Supplemental Fig. S5; Mueller et al. 2011). We validated the specific expression of pvalb7 in larva and adult telencephalon and found strong expression of the gene in the lateral part of the adult telencephalon but weak and spotty expression of the gene in the medial part of the larval telencephalon (Fig. 5C). Together, our ISH and single-cell data indicate that this subtype domain starts to emerge in larva and continues to grow and expand into adulthood.
Conservation and emergence of neuronal diversity between 6-dpf, 15-dpf, and adult telencephalon. (A) The contribution of select pallium clusters to the total pallium cells in 6-dpf, 15-dpf, and adult pallium. (B) Gene expression profiles of pvalb7 faceted by age. (C) In situ expression domain of pvalb7 in larval (green, RNA-FISH; pale gray, total-Erk; costain for anatomy) and adult telencephalon (magenta, RNA-FISH; yellow, Sytox nuclear stain). Closer views of the left and right expression domains in the adult are shown on the corresponding sides of the complete brain section. The scale bars represent 50 µm. (D) The contribution of select subpallium clusters to the total subpallium cells in 6-dpf, 15-dpf, and adult pallium. (E) Gene coexpression profiles for tubb5 with bhlhe22 and six3b. Statistical test for plots in A and D: unpaired two-tailed t-test with Benjamini–Hochberg correction (Benjamini and Hochberg 1995) for multiple comparisons: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 1 × 10−4).
The increase in size of these three pallial clusters was accompanied by a decrease in the proportion of committed pallium precursors and INs in the adult pallium. Among the three IN clusters we identified in the pallium, only Pallium_IN02 and Pallium_IN03 disappear completely in the adult telencephalon. In contrast, Pallium_IN01, which represents the bhlhe22+ class of INs, continues to persist into adulthood despite decreasing in proportion (Fig. 5E, top panel). The three clusters that expand in number belong to this bhlhe22+ cluster of cells, suggesting that these may represent the neuronal cell types that not only expand well into adulthood but also might be capable of regeneration (Kizil et al. 2012; Lange et al. 2020). Indeed, a recent study revealed that vim is up-regulated following telencephalic injury (Kutsia et al. 2022), and vim is a marker of Pallium_IN01 in our data (Fig. 2A, 3A). In addition, the proportion of rprma+ Pallium_02 and pdyn+ Pallium_04 declines steadily from constituting ∼10% of the pallium in larval animals to 2%–3% of the pallium in adults (Fig. 5A).
In contrast, the cellular proportions of the subtypes in the subpallium remain more consistent compared with the pallium. Although the proportions of the committed subpallium precursors and subpallium INs decrease from larva to adult, the only subpallial subtype that expands significantly in size is the six3b+ Subpallium_05 (Fig. 5D). In line with this observation, of the INs that persist in adulthood, a proportion of these is also positive for six3b, suggesting that these INs might be resulting in the expansion of Subpallium_05 later in development (Fig. 5E, bottom panel). These changes in cell proportion of pallium and subpallium neuronal cell types were not a result of disappearing progenitors and INs, as was observed in a separate analysis of cell proportions of just mature neuronal populations (Supplemental Fig. S10A). Finally, although some Subpallium_02 cells persist in adult animals (Fig. 5D), the proportion of the population and corresponding marker genes is substantially reduced (Supplemental Fig. S10B). In addition to the neuronal subtypes, we also found an adult-specific group of endothelial cells (Supplemental Fig. S10C).
Although the key markers of the pallium and subpallium clusters identified by the integrated analysis were largely similar across the 6-dpf, 15-dpf, and adult fish, we performed a systematic comparison of the gene expression profile of each mature neuronal cluster across the three stages analyzed in our study. We generated pseudobulk expression profiles of each pallium and subpallium cluster by separately aggregating cells of different ages into different bulk expression profiles. We then performed cluster-specific DE comparison among all time points: 15 dpf versus 6 dpf (Supplemental Fig. S11A; Supplemental Table S4), adult versus 15 dpf (Supplemental Fig. S11B; Supplemental Table S5), and adult versus 6 dpf (Supplemental Fig. S11C; Supplemental Table S6). Both pallium and subpallium clusters show transcriptional changes despite the identifying markers remaining largely similar across time points. The largest number of DE genes between 15 dpf and 6 dpf as well as between adult and 6 dpf was observed in efna1b+ Pallium_06 (Supplemental Fig. S11A,C), whereas the largest number of DE genes in the 15 dpf versus adult comparison was in pyyb+ Subpallium_02 (Supplemental Fig. S11B).
Integrated analysis is a powerful approach for comparing cell types across related data sets. However, combining the data from the three ages could also mask more subtle differences between the ages. Therefore, we chose to extend our developmental comparison by analyzing the data from each age separately.
Comparison of the larval telencephalon to adult telencephalon with separate clustering
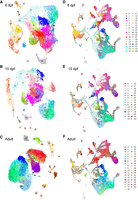
To determine additional similarities and differences between clusters of forebrain cells across developmental stages, we split the data by age and reclustered (Fig. 6A–C). We then mapped each cell to its original location in the integrated space (Fig. 6D–F). The new clusters at each age were connected to the originally defined integrated clusters (Supplemental Table S10) by combining the following information: (1) the location of the cells in relation to the originally defined conserved clusters in the UMAP plot (Fig. 1A), (2) the marker genes for the new clusters at each age (Supplemental Tables S7–S9), and (3) the expression patterns of known marker genes and those identified in the integrated analysis (Supplemental Figs. S12–S17). Clusters that mapped readily to the integrated data are colored identically in Figure 6, whereas new or split clusters are a unique color (Supplemental Table S10).
Separate clustering of each age and the relationship of clusters to the integrated space. (A) UMAP representation of the 6-dpf data clustered separately. (B) UMAP representation of the 15-dpf data clustered separately. (C) UMAP representation of the adult data clustered separately. (D) Visualization of the 6-dpf cells mapped onto the integrated space. (E) Visualization of the 15-dpf cells mapped onto the integrated space. (F) Visualization of the adult cells mapped onto the integrated space.
This separated analysis mirrored some conclusions from the integrated analysis. Similar findings included the loss of the IN types Pallium_IN02 (19 at 6 dpf, 20 at 15 dpf), Pallium_IN03 (18 at 6 dpf, 23 at 15 dpf), and Subpallium_IN (nine at 6 dpf, two at 15 dpf). Pallium_IN01 is present at all three ages but is substantially reduced by the adult stage (clusters 22 and seven at 6 dpf, six and 15 at 15 dpf, 24 at adult). Findings such as the expansion of the Subpallium_05 cluster (21 at 15 dpf, one at adult) and the lack of Pallium_07 cells (not clustered at 6 and 15 dpf, five at adult) until the adult stage are visually apparent when mapping the new clusters in the integrated space (Fig. 6D–F). Other age-dependent changes, such as the reduction in the Pallium_02 and Pallium_04 subtypes (Fig. 5A), are less visually clear but are indicated by the lack of cluster formation (Supplemental Table S10). This difference and others, such as the lack of a cluster forming for Subpallium_01 at any age (Fig. 6D–F; Supplemental Table S10), may be the result of fewer cell numbers per age, leading to a decrease in the power to identify cell types in the separated analyses compared with the integrated.
Although many findings were shared between the two analysis approaches, the separated clustering also identified additional differences between the adult and larval stages. Sample age impacted both neuronal and nonneuronal cell populations. As was recently reported (Wu et al. 2020), there are two types of microglia in the adult zebrafish brain. The adult samples included both the ccl34b.1-expressing phagocytotic microglia and white-matter enriched regulatory microglia that lack ccl34b.1 and express markers such as id2a and batf3, whereas the 6 and 15 dpf only contained the phagocytotic microglia (Supplemental Fig. S18). Second, an immune cell population found in the integrated analysis was only present in the adult ages (Supplemental Fig. S19). We have designated these cells as natural killer (NK)–like, based on a previous study of immune populations in zebrafish that defined top marker genes such as ccl38.5 and il2rb (Carmona et al. 2017). NK cells were recently found in the dentate gyrus of the aging brains of mice and humans (Cuapio and Ljunggren 2021; Jin et al. 2021), supporting this designation. Among the neuronal populations, there were several new clusters formed only in the adult analysis (Fig. 6C), but most had high similarity to identified clusters (Supplemental Table S10). However, one new pallium cluster stood out, characterized by unique marker genes that did not form a cluster in the integrated analysis and were only weakly expressed at larval ages (Supplemental Fig. S20). This cluster, number 26 in the adult data, expresses the marker genes timp2b, dgkzb, rnd3b, and ngef. In mice, TIMP2 is involved in hippocampal-dependent cognition and function and is expressed in the hippocampus (Castellano et al. 2017). It is antimitogenic and promotes neuronal differentiation (Pérez-Martínez and Jaworski 2005). In mammals, DGKZ is expressed in the hippocampus and promotes neurite outgrowth (Kim et al. 2010), as does Rnd3 (Peris et al. 2012; Jie et al. 2015). It is conceivable that this cell population, which is far more prominent in the adult forebrain, is involved in the acquisition of adult-specific learning and memory behavior.
Together, the integrated and separated analysis of these single-cell data sets provides a resource for future investigation into the diversification and transcriptional programming of forebrain cell types as zebrafish develop from larval to adult stages.
Discussion
In this study, we described the molecular architecture of the zebrafish telencephalon at three different stages based on a systematic survey of cell types using single-cell RNA-seq, data integration along the ages, and spatial localization by RNA-FISH. We delineated the identity of these subtypes and their emergence and persistence from larva to adult. The samples spanned (1) an early stage (6 dpf) when the larval behavioral repertoire is not thought to include complex behaviors such as learning or social behavior, (2) a middle stage (15 dpf) when complex behavioral repertoires begin to emerge (Valente et al. 2012; Dreosti et al. 2015; Palumbo et al. 2020; Stednitz and Washbourne 2020) and (3) an adult stage when the telencephalon is fully formed and complex behaviors such as learning and memory are well established (Aoki et al. 2013; Cheng et al. 2014; Lal et al. 2018; Stednitz et al. 2018). Furthermore, we integrated our single-cell expression data set from zebrafish with a previously published mouse data set to delineate the putative relationship between zebrafish and mammalian neuronal cell types. These single-cell data, together with the RNA-FISH-based anatomical map (Supplemental Video S1), will be an important resource for the dissection of function and development of the zebrafish telencephalon.
A comparison of the zebrafish and mouse telencephalon revealed both conserved and divergent cell types. Although the pallium is one of the most evolutionarily and structurally divergent areas of the vertebrate brain, computational integration of mouse and zebrafish single-cell RNA-seq data (Fig. 4C,D) clustered zebrafish types with mammalian brain regions that were previously proposed (e.g., Pallium_08 cells in the cortical-like area) (Ganz et al. 2014). Multiple zebrafish subpallium clusters integrated with the expected mouse cell types (e.g., Subpallium_01, Subpallium_06). The computational integration was not equally successful for all clusters. For example, Pallium_01 and Pallium_04 both cluster near the basolateral amygdala and anterior olfactory neurons (Fig. 4C,D), unrelated to their expected cell types from anatomical analysis (Ganz et al. 2014; Porter and Mueller 2020), and do not share obvious marker genes with single mouse clusters. The Pallium_01 cluster is marked by pyya, and a corresponding population of Pyy+ neurons in the murine telencephalon has not been identified. Subpallium_04 also clusters near the basolateral amygdala and inhibitory interneurons, but there is no obvious cluster corresponding to glutamatergic septal cells in the adolescent mouse data set. The absence of this mouse cluster may be technical, as it is an evolutionarily conserved cell type (Woych et al. 2022).
Differences in marker gene expression between the species may represent a true divergence or a technical outcome of RNA or cell type loss. Some classical mammalian cortical markers, such as satb2, are absent from the zebrafish pallium and expressed only in the subpallium, consistent with previous reports (Furlan et al. 2017; Lozano et al. 2022). This discrepancy likely represents a functional difference, as this factor has been implicated in the evolution of the neocortex and cortical neurogenesis (Otani et al. 2016; Lozano et al. 2022). The zebrafish correlate of Vip+/Calb2+ interneurons is also missing, which is surprising given previous reports of calretinin in the zebrafish telencephalon (Castro et al. 2006; Kenney et al. 2021). It has been reported that some cell types, including interneurons, are undersampled in single-cell RNA-seq because of their fragility (Zeisel et al. 2018). In other clusters, the transcriptional subtype appears conserved but is missing a few expected marker genes. For example, although Subpallium_02 and Subpallium_05 express many markers of D1 and D2 MSNs, respectively, neither express substantial dopamine receptor transcripts in our data set. Orthogonal approaches, such as spatial transcriptomics, will be required to clarify whether these differences are biological or technical.
We compared the larval and adult telencephalon samples in two ways: using the integrated analysis to observe how conserved clusters changed with age and using a separated analysis to uncover additional clusters. The two approaches yielded both complementary and unique information. The integrated analysis revealed how populations expressing conserved markers expanded and shrank during forebrain maturation. The shrinking of mature neuron clusters is likely owing to these neuron types staying constant in number whereas others expand or to changes in their marker gene expression. The separated analysis revealed both neuronal and nonneuronal cell types that were only present at the adult stage. Some align with previously reported results, such as the adult-specific population of regulatory microglia (Supplemental Fig. S18; Wu et al. 2020), affirming the robustness of our atlas. Other findings are novel, such as the adult-specific endothelial cells (Supplemental Fig. S10C) and the presence of NK-like cells in the zebrafish forebrain (Supplemental Fig. S19). In future work, as the number of cells sampled in these brain regions increases, further subclustering of heterogenous clusters such as Subpallium_07 (Supplemental Fig. S5) is likely to reveal additional new cell types.
Both the integrated and separated comparative analyses showed that the telencephalon, particularly the pallium, grows not just in size during development from larva to adult but also selectively expands certain neuronal cell types that may be essential for the emergence of complex behaviors such as learning in adulthood. For example, the Pallium_07 cluster expands substantially between the larval and adult stages (Fig. 5). Parvalbumin neurons in the pallium have been shown to be important for the retrieval in aversive reinforcement learning, a behavior that is only displayed by fish at or after the juvenile stages (Aoki et al. 2013). Similarly, cells expressing the top markers of the adult pallium cluster 26 (timp2b, dgkzb, rnd3b, ngef) were only found in the mature brain (Supplemental Fig. S20), and these markers are associated with hippocampal formation and function in mammals. Beyond these specific clusters, it is not clear whether the additional apparent complexity in the pallium (Fig. 6C; Supplemental Table S10) is owing to differences in clustering or represents true differences between the larval and mature brain. Although changes were also observed in the subpallium, the ratios of the conserved neuronal subtypes remain more consistent, suggesting that it may generate distinct neuronal cell types earlier in development than the pallium.
Although there have been several large-scale single-cell sequencing studies of zebrafish in recent years, we provide an in-depth investigation of the telencephalon at three stages of brain development. A focused regional analysis of individual brain regions can uncover cell types that are not revealed with more shallow analyses of the entire animal or brain (Pandey et al. 2018). The telencephalon undergoes massive growth, neurogenesis, and functional maturation from the developing larval to mature adult stage, and few studies have explored its cellular complexity at adult stages. As several behaviors mediated by neuronal ensembles in the telencephalon emerge only in adults, such as social behaviors and fear learning, this atlas provides new cluster-specific marker genes to bolster future studies of the relationship between neuron types and complex behaviors.
Methods
Experimental model
Wild-type larvae and adult fish of the TLAB strain were maintained on a 14-h:10-h light–dark cycle at 28°C. All protocols and procedures involving zebrafish were approved by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research and Teaching (IACUC; protocol 25-08); 6-dpf, 15-dpf larval, and ∼1-yr-old adult zebrafish were used. The four adult samples consisted of two female (b1 samples) and two male (b2 samples) brains. Animals were anesthetized in 0.2% tricaine and rapidly euthanized by immersion in ice water for 5 min before dissection.
Cell isolation and single-cell RNA-seq
Forebrains were dissected from wild-type fish in Neurobasal (Thermo Fisher Scientific 21103049) supplemented with 1× B27 (Thermo Fisher Scientific 17504044) and promptly dissociated with the papain dissociation kit (Worthington Biochemical Corporation LK003150). Per sample, six to eight forebrains were pooled for 6 dpf, two for 15 dpf, and two for adult. For the adult samples, two 10x wells were used, for a total of four final samples. Larval forebrains were incubated in 20 units/mL papain for 12 min at 37°C, 15 dpf for 16 min, and adult for 30 min. The cells were dissociated by gentle trituration 20 times and spun at 300g for 5 min. The cells were resuspended in 1.1 mg/mL papain inhibitor in Earle's balanced salt solution (EBSS) and spun at 300g for 5 min. The resulting pellet was then washed in neurobasal supplemented with B27 before final resuspension in 50 µL PBS + 200 µg/mL BSA. To avoid cell loss during each wash, the supernatant was saved and spun a second time to recover the remaining cells. The cells were then mixed with the final suspension to increase cell count. Viability and cell number were assessed by trypan blue staining on 10 µL of the sample. If viability was >80%, the cells were loaded on the 10x Chromium system at a concentration of approximately 150 cells per microliter. Two samples were generated for each of the adult forebrains. Libraries were prepared on the 10x Genomics scRNA-seq platform according to the manufacturer's instructions (single-cell 3′ v2 kit). Single-cell transcriptome libraries were sequenced using Nextera 75 cycle kits at the Bauer Core Facility (Harvard).
Fluorescent RNA in situ hybridization
Fluorescent RNA in situ hybridizations were performed as previously described (Ronneberger et al. 2012; Pandey et al. 2018) using 10 dpf mitfa−/− larvae or adult samples. We chose 10 dpf for efficient whole-mount data collection that would provide spatial information relating to two of our time points (6 and 15 dpf), as well as for consistency with our published work on the habenula (Pandey et al. 2018). To generate probes, gene fragments were amplified from cDNA with Phusion polymerase (New England Biolabs M0530L) using the primers that are listed in Supplemental Table S2. The polymerase chain reaction (PCR)–amplified fragments were then cloned into pSC-A plasmid using the StrataClone PCR cloning kit (Agilent 240205) and StrataClone-competent cells. The transformed cells were plated overnight on Luria–Bertani (LB) agar plates. Colonies were selected by colony PCR, cultured, mini-prepped, and sent for sequencing. The resulting plasmids were then restricted with the appropriate restriction enzyme and purified using a PCR-clean up kit (Omega cycle-pure kit). The linearized vector was then used as a template to synthesize digoxigenin- or fluorescein-labeled RNA probes using the RNA labeling kit (Roche). The transcription reactions were purified using a total RNA clean-up kit (Omega R6834), and the resulting RNA was quantified using NanoDrop and assessed on an agarose gel. The final product was then normalized to 50 ng/μL in HM+ buffer (50% formamide, 5× saline sodium citrate [SSC] buffer, 5 mg/mL torula RNA, 50 μg/mL heparin, 0.1% Tween 20) and stored at −20°C until further use. Adult samples were processed using the preceding protocol with the following modifications: Dissected brains were digested in 20 µg/mL Proteinase K for 35 min, mounted in 3% low-melt agarose, sliced into 50-µm sections using a vibratome, stained with Sytox green (1:30,000), and imaged using an inverted Zeiss LSM 880 confocal with a 20× air objective and a 63× oil dipping objective.
Subsequent immunostaining of larval samples with the anti-total-Erk p44/42 MAPK (Erk1/2) antibody (Cell Signaling Technology 9102) was performed as previously described (Randlett et al. 2015). The use of the total-Erk counterstain facilitates anatomical image registration, similar to the use of Z-Brain atlas to compare three-dimensional stacks (Randlett et al. 2015). Larvae were washed three times in PBST, mounted in 2% low-melt agarose, and imaged with an upright confocal Zeiss LSM 880 using a water-dipping 20× objective. Imaging data were typically collected with a width × height of 1024 × 1024 and imaging resolution of 3.6 pixels per micrometer. To generate Supplemental Video S1, images were registered using CMTK (Rohlfing and Maurer 2003), as previously described (Pandey et al. 2018).
Computational methods for data analysis
All single-cell RNA-seq analyses were performed using R Statistical Software (R Core Team 2021) and are available as HTML files at GitHub (https://github.com/sthyme/scrnaseq-zfforebrain).
Alignment, quantification, and filtering for single-cell data sets
Raw sequencing data were converted to matrices of expression counts using the Cell Ranger software provided by 10x Genomics. Briefly, raw BCL files from the Illumina NextSeq or HiSeq were demultiplexed into paired-end, gzip-compressed FASTQ files for each channel using “cellranger mkfastq.” Both pairs of FASTQ files were then provided as input to “cellranger count,” which partitioned the reads into their cell of origin based on the 14-bp cell barcode on the left read. Reads were aligned to a zebrafish reference transcriptome (Ensembl Zv10, release 82 reference transcriptome), and transcript counts were quantified for each annotated gene within every cell. Here, the 10-bp unique molecular identifier (UMI) on the left read was used to collapse PCR duplicates and accurately quantify the number of transcript molecules captured for each gene in every cell. Both Cell Ranger mkfastq and Cell Ranger count were run with default command-line options. This resulted in an expression matrix (genes × cells) of UMI counts for each sample. Of the nine samples from 6-dpf larvae, one (b0) (Supplemental Fig. S2) was previously published and can be obtained from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE115427 (Thyme et al. 2019). The Zv10-based transcriptome is compatible with previous zebrafish single-cell neuronal data sets (Pandey et al. 2018; Raj et al. 2020). This transcriptome mapping contains two unpublished genes not found in the Ensembl Zv10: NP5 (Chr 16: 20,355,986–20,358,635), now known as c16h2orf66, and NP33 (Chr 16: 47,449,630–47,452,948). To compare the results of mapping to Zv10 and the more recent Zv11 genome, reads from the 15-dpf samples were aligned using the newest, most comprehensive published zebrafish transcriptome data (Lawson et al. 2020) and STARsolo (Kaminow et al. 2021; Brüning et al. 2022). The final marker genes for clusters in the newly aligned data (Supplemental Table S11) were defined by comparable Seurat clustering as was completed for the original 15-dpf data set (Supplemental Table S8).
Data set integration and clustering analysis
Using the expression matrix, cells were filtered to remove those that contained fewer than 200 genes and those in which >8% of transcript counts were derived from mitochondrial-encoded genes. To select highly variable genes, we used a union of a UMI-based method described recently (Pandey et al. 2018) and Seurat's (Satija et al. 2015) variable gene selection approach. To correct for batch effects as well as to identify shared and stage-specific clusters, we used Seurat's data integration methodology that combines CCA and MNN (Seurat v 3.2.2). Seurat uses CCA to identify correlated gene expression changes in the cells between the stages and then identifies and maps the clusters that are most similar across the different stages. These genes and the respective corrected expression values were then used for dimensionality reduction and clustering analysis. Variable genes from all three stages were used for the integration process. The data were clustered using the high resolution of two, yielding 51 clusters, and then these clusters were merged based on shared markers and a lack of unique markers to reach 38 clusters. The analysis of the separate ages was performed by subsetting each age from the integrated object and reclustering. The clustering resolution chosen for each age was guided by the clustree package (Zappia and Oshlack 2018) and an examination of the cluster marker genes. Although there may be overclustering of some pallial subtypes in the adult data at the chosen resolution of 1.6 (Supplemental Table S10), reducing the resolution further resulted in the loss of clusters that were expected to remain distinct.
Marker gene identification
To annotate clusters and identify genes that are enriched within each cluster, we used Seurat's FindMarker function using the “Wilcoxon” test. Uncorrected expression data (DefaultAssay(object) ← “RNA”) was used for this purpose. This allowed the identification of stage-specific marker genes. To identify conserved markers between the stages, we used Seurat's FindConservedMarkers function.
Identification of DE genes between 6-dpf, 15-dpf, and adult pallium and subpallium clusters
To identify the genes that are DE between the different ages analyzed in our study, we generated pseudobulk profiles of all mature neuronal clusters from the pallium and subpallium. Pseudobulk expression profiles were derived from single-cell data sets by aggregating cells of a given sample of the same cell type separately such that a total of sc possible pseudobulks were generated for s samples and c cell types. If a particular pseudobulk contained fewer than 50 cells, these profiles were discarded such that the actual number of pseudobulks was always less than the theoretical maximum number of pseudobulks, sc. For each pseudobulk profile, raw counts were generated by adding the total number of UMIs for each gene across all the cells belonging to a particular sample and cell type. This resulted in a gene-by-pseudobulk count matrix, which was normalized and used for differential gene expression analysis using edgeR (Robinson et al. 2010). These normalized pseudobulk profiles were then used for differential gene expression analysis between groups of interest using UpSet plots (Supplemental Fig. S11; Lex et al. 2014; Conway et al. 2017). All DE analysis was performed with voom (Law et al. 2014), and genes were filtered as differentially expressed if the log fold change was greater than or equal to two and the adjusted P-value was ≤0.05.
Pseudotime analysis
To construct a single-cell trajectory, Committed_pallium_precursors, Pallium_IN01, Pallium_IN02, Pallium_02, Pallium_03, Pallium_05, Pallium_06, and Pallium_07 were reclustered using Monocle 3 (version monocle3_1.3.1) with default parameters (Trapnell et al. 2014; Cao et al. 2019). After fitting a principal graph, the committed pallium precursors were specified as the root node. Modules of genes that vary as a function of pseudotime were identified with a clustering resolution of 5 × 10−3.
Integration of zebrafish and mouse data
Zebrafish gene names for cells in mature neuronal clusters (Pallium_01-09 and Subpallium_01-08) were converted to mouse gene symbols using the Orthogene R package (version orthogene_1.4.1) with “keep popular” mapping option (https://doi.org/doi:10.18129/B9.bioc.orthogene). Gene counts for zebrafish genes with identical mouse mappings were averaged. Pvalb, which has zebrafish homologs expressed in distinct clusters (pvalb6 and pvalb7), was removed from the analysis. Forebrain neuronal clusters from adolescent mouse (Zeisel et al. 2018) were downloaded from http://mousebrain.org/adolescent/downloads.html, imported using loomR (version loomR_0.2.0), and converted to “Seurat” object using the as.Seurat function. Mouse clusters included in the analysis were TEGLU1, TEGLU3, TEGLU2, TEGLU20, TEGLU11, TEGLU12, TEGLU10, TEGLU9, TEGLU8, TEGLU7, TEGLU6, TEGLU13, TEGLU14, TEGLU5, TEGLU16, TEGLU15, TEGLU17, TEGLU18, TEGLU19, TEGLU22, TEGLU21, TEGLU4, TEGLU24, TEGLU23, CR, DECHO1, MSN1, MSN2, MSN3, MSN4, MSN5, MSN6, TEINH17, TEINH18, TEINH19, TEINH21, TEINH16, TEINH15, TEINH14, TEINH20, TEINH13, TEINH12, TEINH9, TEINH10, TEINH11, TEINH4, TEINH5, TEINH8, TEINH7, TEINH6, TECHO, TEINH3, TEINH2, and TEINH1. A total of 11,843 genes across 68,231 cells were included in the analysis.
For integrated zebrafish and mouse analysis, cells with fewer than 200 genes and cells with >6% mitochondrial transcripts were removed. UMI counts were normalized using SCTransform (Hafemeister and Satija 2019) with the percentage of mitochondrial transcripts used as a variable for regression. Harmony (version harmony_0.1.1) (Korsunsky et al. 2019) was used to integrate mouse and zebrafish cells, followed by standard Seurat analysis using the first 30 principal components and a clustering resolution of 0.35. Cells were colored according to their original cluster identity in zebrafish or probable location in the mouse (Zeisel et al. 2018).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE212314. The final objects are available as an R Shiny application on Docker, along with instructions for deployment at GitHub (https://github.com/U-BDS/zebrafish_telencephalon_atlas) and as Supplemental Code. The R Shiny application is also hosted at https://zfforebrain.thymelab.org/.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank Alexander Schier for supporting this study and his funding sources of the National Institutes of Health (NIH) DP1HD094764, Allen Discovery Center grant, and McKnight Foundation Technological Innovations in Neuroscience Award. This work was also supported by NIH grant R00MH110603 and a Klingenstein-Simons Award in Neuroscience to S.B.T. and T32HG008961 to A.J.M. We acknowledge support in the development of the R Shiny app from Lara Ianov and the University of Alabama at Birmingham Biological Data Science Core, RRID:SCR_021766. We thank Bushra Raj, Maxwell Shafer, and Mahima Reddy for helpful comments on the project; the Bauer Core Facility (Harvard) for sequencing services; the UAB Research Computing team for the Cheaha computer cluster; the Harvard zebrafish facility staff for technical support; and the Harvard Center for Biological Imaging for microscopy resources.
Author contributions: S.P. designed the study and performed the collection of the single-cell data and the fluorescent in situ hybridization data. S.B.T. cloned constructs for generating ISH probes and generated the registered map of the telencephalon. S.P., A.J.M., and S.B.T. analyzed the data and wrote the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277278.122.
- Received September 2, 2022.
- Accepted April 11, 2023.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see https://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.