Regulatory Roles of Conserved Intergenic Domains in Vertebrate Dlx Bigene Clusters
- Noël Ghanem1,2,7,
- Olga Jarinova1,2,7,
- Angel Amores3,
- Qiaoming Long1,2,5,
- Gary Hatch1,
- Byung Keon Park1,6,
- John L.R. Rubenstein4, and
- Marc Ekker1,2,8
- 1Ottawa Health Research Institute and 2Department of Cellular and Molecular Medicine, University of Ottawa, Ottawa, Ontario, Canada K1Y 4E9; 3Institute of Neuroscience, University of Oregon, Eugene, Oregon 97403, USA; 4Nina Ireland Laboratory of Developmental Neurobiology, Center for Neurobiology and Psychiatry, Department of Psychiatry and Programs in Neuroscience, Developmental Biology and Biomedical Sciences, University of California at San Francisco, California 94143–0984, USA.
Abstract
Dlx homeobox genes of vertebrates are generally arranged as three bigene clusters on distinct chromosomes. The Dlx1/Dlx2,Dlx5/Dlx6, and Dlx3/Dlx7 clusters likely originate from duplications of an ancestral Dlx gene pair. Overlaps in expression are often observed between genes from the different clusters. To determine if the overlaps are a result of the conservation of enhancer sequences between paralogous clusters, we compared theDlx1/2 and the Dlx5/Dlx6 intergenic regions from human, mouse, zebrafish, and from two pufferfish, Spheroides nephelus and Takifugu rubripes. Conservation between all five vertebrates is limited to four sequences, two inDlx1/Dlx2 and two in Dlx5/Dlx6. These noncoding sequences are >75% identical over a few hundred base pairs, even in distant vertebrates. However, when compared to each other, the four intergenic sequences show a much more limited similarity. Each intergenic sequence acts as an enhancer when tested in transgenic animals. Three of them are active in the forebrain with overlapping patterns despite their limited sequence similarity. The lack of sequence similarity between paralogous intergenic regions and the high degree of sequence conservation of orthologous enhancers suggest a rapid divergence of Dlx intergenic regions early in chordate/vertebrate evolution followed by fixation ofcis-acting regulatory elements.
[Supplemental material is available online at www.genome.org.]
Vertebrates possess anatomical features not seen in their closest living invertebrate relatives, the protochordates such as tunicates and cephalochordates. Genetic changes, such as the evolution of new regulatory pathways, may have permitted the origin of these innovations. Gene duplication followed by functional divergence of paralogs constitutes a major mechanism that permits such changes. An important contribution to the evolutionary divergence of paralogs may be through changes in mechanisms that control gene expression viacis-acting regulatory sequences in the noncoding region of genes. However, the identification of cis-acting regulatory elements remains challenging, even after the completion of a few vertebrate genome sequences.
The vertebrate Dlx genes, which encode a family of homeobox-containing transcription factors related in sequence to theDrosophila Distal-less (Dll) gene product, constitute one example of functional diversification of paralogs. All vertebrates investigated thus far have at least six Dlx genes that are generally arranged as three bigene clusters: Dlx1/Dlx2,Dlx5/Dlx6, and Dlx3/Dlx7 (Simeone et al. 1994;McGuinness et al. 1996; Nakamura et al. 1996; Stock et al. 1996; Ellies et al. 1997; Liu et al. 1997). Each bigene cluster is localized on a distinct chromosome that also contains one of the Hoxclusters, suggesting that the duplication events that generated the multiple Dlx bigene clusters of vertebrates also involved theHox genes (Stock et al. 1996; Amores et al. 1998). The two linked Dlx genes are in an inverted configuration and separated by a short intergenic (3.5–16 kb) region. Because only oneDll-like gene is found in invertebrates such asDrosophila and Caenorhabditis elegans, the multiple vertebrate Dlx genes are thought to have arisen as a result of tandem gene duplication events from one “hypothetical” common ancestor to nematodes, arthropods, and vertebrates. The presence, in the tunicate Ciona intestinalis of pair of Dll-like gene with an organization similar to that of the vertebrateDlx (Di Gregorio et al. 1995; Caracciolo et al. 2000) supports the hypothesis that the initial duplication predated the existence of vertebrates.
Gene families such as the Dlx family provide attractive models for studying gene regulation and functional divergence between paralogs. The bigene cluster arrangement of Dlx genes is conserved amongst distant vertebrates and a direct association is seen between the genomic organization of the genes and their expression pattern in different species (Ellies et al. 1997; Zerucha et al. 2000) suggesting that the mechanisms of regulation might have been conserved, at least in part. Functional conservation among different orthologs, as inferred from comparative expression patterns seems to be applicable to most vertebrate Dlx genes (Quint et al. 2000; Zerucha and Ekker 2000). Partial functional redundancy between Dlxparalogs is suggested by the overlapping gene expression patterns and phenotypes of mice with targeted Dlx mutations (Qiu et al. 1995, 1997; Anderson et al. 1997; Acampora et al. 1999; Depew et al. 1999; Robledo et al. 2002). Sharing of cis-regulatory elements between members of a Dlx bigene cluster may contribute to the overlap in gene expression and to their partial functional redundancy.
Consistent with a model of enhancer-sharing, two highly conserved enhancer elements, I56i and I56ii, were identified in the intergenic region of the Dlx5/Dlx6 genes of zebrafish, mouse, and human and were able to target expression of reporter transgenes to the forebrain of both mouse and zebrafish in patterns that mimic the endogenous gene expression (Zerucha et al. 2000). Recently, Sumiyama and collaborators conducted a comparative sequence analysis of the mouse and human Dlx3/Dlx7 (Dlx3/Dlx4 was suggested as revised nomenclature by Panganiban and Rubenstein 2002) bi-gene cluster (Sumiyama et al. 2002). Conserved sequences were identified both in the coding and noncoding regions of Dlx3/Dlx7. Comparisons of the two mammalian loci with the orthologousdlx3/dlx7 bigene cluster from zebrafish revealed a much more limited similarity (Sumiyama et al. 2002).
The two genes from the Dlx1/Dlx2 cluster are expressed in the developing forebrain with patterns that overlap partially with those ofDlx5 and Dlx6. As the Dlx1/Dlx2 andDlx5/Dlx6 bigene clusters probably originate from the duplication of an ancestral cluster, the forebrain expression ofDlx1 and Dlx2 could be attributable to enhancer sequences related to I56i and/or I56ii. To address this possibility and to get a comprehensive understanding of cis-acting regulatory elements in the Dlx1/Dlx2 and Dlx5/Dlx6 intergenic regions, we have performed a homology search (phylogenetic footprinting) between the intergenic regions of the two bigene clusters from five vertebrate species: human, mouse, zebrafish, Takifugu rubripes (formerly Fugu rubripes) and Spheroides nephelus. Sequence conservation between all five species is limited to four distinct sequences of a few hundred base pairs, two in each intergenic region. Each sequence shows enhancer activity in transgenic mice and/or zebrafish. A novel forebrain enhancer, I12b, was identified in the Dlx1/Dlx2 intergenic region, but surprisingly, it shows almost no sequence similarity to the I56i and I56ii forebrain enhancers, suggesting that highly overlapping patterns of expression can be conferred by highly different cis-acting regulatory sequences.
RESULTS
Genomic Organization of Dlx1/Dlx2 and Dlx5/Dlx6Bigene Clusters in Two Species of Pufferfish
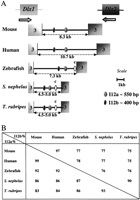
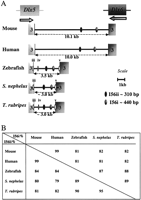
The genomic organization of two loci containing Dlx genes was examined in Spheroides nephelus and Takifugu rubripes and was compared to that of zebrafish, mouse, and human. Initial orthology assignment was based on the sequence of the third exon of the genes, which contains part of the homeobox. Orthology was further confirmed by sequence analysis of the intergenic region. As previously described for zebrafish, mouse, and human (Simeone et al. 1994; McGuinness et al. 1996; Ellies et al. 1997; Zerucha et al. 2000), the dlx1/dlx2 genes and the dlx5/dlx6 genes ofSpheroides and Takifugu are organized as two pairs of genes, both found in an inverted and convergent configuration (Figs.1A, 2A).
Conserved sequences in the Dlx1/Dlx2 intergenic region. (A) Schematic representation of the Dlx1/Dlx2intergenic region of five vertebrate species. The third exons of theDlx genes are indicated. The position of the polyadenylation sequence in the Dlx genes of Spheroides andTakifugu is an estimate. In addition to the I12a and I12b sequences, ovals labeled “c” represent a region of sequence conservation between the three teleost fish species. (B) Percentage identity for I12a and I12b in pairwise sequence comparisons.
Conserved sequences in the Dlx5/Dlx6 intergenic region. (A) Schematic representation of the Dlx5/Dlx6intergenic region of five vertebrate species. The third exon of theDlx genes are indicated. The position of the polyadenylation sequence in the Dlx genes of Spheroides andTakifugu is an estimate. In addition to the I56i and I56ii sequences, ovals labeled iii, iv, and v represent regions of sequence conservation between a subset of the five species. Sequence alignments can be found as supplemental files. (B) Percentage identity for I56i and I56ii in pairwise sequence comparisons.
The size of the Dlx1/Dlx2 intergenic region in the five species varies between about 4.5–5.0 kb for the two pufferfish to 10.7 kb for human (Fig. 1A). It was difficult to determine with precision the size of the pufferfish intergenic regions because no cDNA sequences are available for the Dlx1 and Dlx2 genes from these species and unequivocal polyadenylation signals were sometimes hard to find in the genomic sequence. The distance that separates the two stop codons is 5.3 kb in both species.
The size of the Dlx5/Dlx6 intergenic region varied between 10 kb for mouse and human and about 3.0–3.5 kb for the three teleost fish (Fig. 2A). Thus despite the fact that the genome size for Takifugu rubripes and Spheroides nephelus is ∼4 and 8 times smaller than those of the zebrafish and mouse/human, respectively, this is not reflected in proportionally smaller intergenic regions.
Sequence Comparisons and Identification of Highly Conserved Noncoding Sequence Elements in the Dlx Intergenic Regions
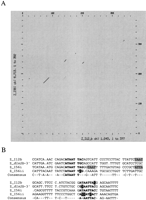
We examined the Dlx1/Dlx2 and Dlx5/Dlx6 intergenic regions of the five vertebrate species for conserved sequences. The mouse and human Dlx1/Dlx2 intergenic regions were highly similar with 80% overall sequence identity (Fig.3A). The same applies for the humanDlx5/Dlx6 intergenic region (78% Fig. 3B) and for thedlx1/dlx2 and dlx5/dlx6 intergenic regions ofTakifugu rubripes and Spheroides nephelus with 85% and 87% sequence identity, respectively (data not shown). This reflects the relatively recent divergence from one common ancestor between mouse and human (∼60 million years), on the one hand, and between the two species of pufferfish, on the other hand (between 5–35 million years). Despite the high degree of sequence conservation between orthologous loci, the paralogous intergenic regions,Dlx1/Dlx2 and Dlx5/Dlx6, do not show any striking sequence similarity and no large regions of sequence similarity can be found between the intergenic sequence separating Dlx3 andDlx7 of human, mouse, and zebrafish (Sumiyama et al. 2002).
Percentage identity plot (PIP) of the (A) Dlx1/2, and (B) Dlx5/6 intergenic regions between mouse, human, and zebrafish. The mouse sequence is shown on the horizontal axis and the percentage identity to the human (top plot) and zebrafish sequences (lower plot) are shown on the vertical axis. Sequences used for comparison include the intergenic regions and the 3′UTRs of both flanking genes. In A, Dlx1 is to theleft and in B, Dlx5 is to the left. Shaded dark and light gray areas indicate the positions of enhancers. Repetitive sequences are shown as follows: black triangles, mammalian interspersed repeats (MIR); vertical rectangles, simple sequence repeats; CpG islands: white horizontal rectangle, CpG ratio >0.60; gray rectangles, CpG ratio >0.75. For further details on PIP analyses, see http://bio.cse.psu.edu/pipmaker.
Two highly conserved sequences that were previously identified in theDlx5/Dlx6 intergenic region of zebrafish, mouse, and human (Zerucha et al. 2000), I56i and I56ii, were also found in thedlx5/dlx6 intergenic regions of Takifugu andSpheroides. They constitute the only two regions of high sequence similarity between all five species (Fig. 2A, 3B). The sizes of I56i and I56ii are ∼440 bp and 310 bp, respectively, and the identity percentages in pairwise comparisons vary between 81 and 99% (Fig. 2B; five-species alignment provided as supplementary Figs. 1 and 2). The relative positions and orientation of the I56i and I56ii sequences with respect to the flanking genes were identical for all five vertebrates. In both the mouse/human (Fig. 3B) and theTakifugu/Spheroides (not shown) alignments, I56i and I56ii reside in a region of overall stronger sequence conservation.
In addition to I56i and I56ii, we found two sequences of 150–200 bp with >80% identity between zebrafish, Takifugu, andSpheroides (Fig. 2A; alignments provided as supplementary Figs. 3 and 4). The first is found in the 3′UTR sequence of zebrafishdlx5a (see note concerning the nomenclature of zebrafishdlx genes in the Methods section) and at a corresponding position, with respect to the predicted stop codons of theTakifugu and Spheroides orthologs (Fig. 2A). The second is found just downstream of the 3′UTR of zebrafishdlx6a and at a similar position in the pufferfish orthologs. Finally, a fragment of about 100 bp with 83% sequence identity was found between the end of dlx5a and I56ii in zebrafish andTakifugu but was not found in Spheroides (alignment provided as supplementary Fig. 5). None of the three shorter conserved sequences could be identified in the two mammalian loci.
We identified two highly conserved sequences in the Dlx1/Dlx2intergenic regions of the five vertebrates. The first, I12a, is ∼550 bp in length and the percentages in sequence identity in pairwise comparisons vary between 83% and 99% (Figs. 1B,4). The second, I12b, is about 400 bp in length and shows percentages of identity that vary between 75% and 97% (Figs. 1B, 5). The relative positions and orientations of I12a and I12b with respect to the Dlx1 andDlx2 genes were identical in all five species. As for I56i and I56ii, the I12a and I12b sequences reside in a region of overall stronger sequence conservation in mouse/human (Fig. 3A) and inTakifugu/Spheroides (not shown) pairwise comparisons.
Multiple sequence alignment of I12a in five vertebrate species; M, mouse; H, human, T, Takifugu rubripes; S, Spheroides nephelus; and Z, zebrafish. The consensus sequence represents identity in four out of five species.
Multiple sequence alignment of I12b in five vertebrate species. The consensus sequence represents identity in four out of five species. Sequences similar to the binding site for Dlx protein ([A/C/G]TAATT[G/A][C/G]) (Feledy et al. 1999) are shown in bold. See also Figure 7. Abbreviations as in Figure 4.
In addition to I12a and I12b, we identified a sequence of ∼320 bp, I12c, that was conserved between Takifugu,Spheroides, and zebrafish. This sequence is located between the end of dlx2 and I12a (Fig. 1A; alignment provided as supplementary Fig. 6). Finally, a sequence of ∼110 bp was found in or near the 3′UTR of Dlx1 of mouse and human and the in the zebrafish dlx1/dlx2 locus, between the 3′end of dlx1and I12b (alignment provided as supplementary Fig. 7). This sequence contains a TTA tri-nucleotide repeat but sequence conservation extends beyond this repeat.
The Sequences Conserved Between All Five Vertebrate Species Contain Enhancers
To determine that the conserved Dlx intergenic sequences, I56i, I56ii, I12a, and I12b, constitute cis-acting regulatory sequences, they were tested in reporter constructs that were injected to produce transgenic mice and zebrafish. As previously reported, I56i and I56ii target expression of lacZ reporter constructs to the forebrain of transgenic mice and zebrafish starting at E10 and persisting in adult mice (Zerucha et al. 2000). The mouse I56i sequence can efficiently target expression to the forebrain by itself in 100% of primary transgenic mice expressing the transgene and in three out of four transgenic lines (Fig. 6A; Table1) (Zerucha et al. 2000). The zebrafish I56i sequence also targeted expression to the forebrain of 12 out of 12 primary transgenic mouse embryos (Zerucha et al. 2000). In both cases, reporter gene expression precisely mimics that of the endogenous Dlx5 gene and highly overlaps with that of Dlx6 (Zerucha et al. 2000).
Enhancer activity of conserved Dlx intergenic sequences in transgenic mice (A–E) and zebrafish (F–J). (A) Mouse I56i, (B) mouse I56ii, and (C) mouse I12b each drive reporter gene expression to the telencephalon (BT) and diencephalon (Di) of transgenic mice, as shown here in mouse embryos. (D, E) Mouse I12a drives reporter gene expression to a subset of mesenchymal cells in the mandibular (Md) component of the first branchial arch and in the second branchial arch (Hy) of an E11.5 embryo. (A–D) are sagittal views and (E) is a frontal view of the embryo shown in (D). All embryos are at stage E11–12. FN, frontonasal prominence. (F) Head of 48 hpf primary transgenic zebrafish embryo, dorso-lateral view, injected with the controldlx6a-GFP reporter plasmid. Injection of this construct results in very few GFP-positive cells with no tissue specificity (n > 150). (G,H) Lateral and frontal views, respectively, of a 48-hpf zebrafish embryo from a transgenic line produced with a construct made with the dlx6a-GFP reporter plasmid that also contained a 1.4-kb dlx5a/dlx6a intergenic fragment containing I56i and I56 ii. I and II indicate the diencephalic and telencephalic domains of transgene expression, which also correspond to endogenous dlx expression patterns in the zebrafish forebrain. (I) Frontal view of a 48-hpf primary transgenic zebrafish embryo injected with a dlx6a-GFP that also contained a 4.0-kb mouse Dlx5/Dlx6 intergenic fragment that comprises I56i. The transgene is expressed predominantly in the telencephalic domain II. (J) Lateral view of a 48-hpf primary transgenic zebrafish embryo injected with a dlx6a-GFP that also contained a 2.8-kb mouse Dlx5/Dlx6 intergenic fragment that comprises both I56i and I56ii. GFP-positive cells are seen only in the telencephalic domain, II.
Expression of Reporter Constructs in Primary Transgenic Mouse Embryos and Transgenic Mouse Line
Three primary transgenic mice and two established lines containing a mouse I56ii reporter construct expressed lacZ in the forebrain (Fig. 6B), although the intensity of the ß-galactosidase staining was more variable between the telencephalic and diencephalic expression domains, and staining seemed often weaker than that observed with I56i constructs. However, the mouse I56ii (this work) was more efficient at targeting transgene expression to the forebrain than its zebrafish counterpart (Zerucha et al. 2000).
When tested in transgenic zebrafish, a construct containing both zebrafish I56i and I56ii targeted expression of the green fluorescent protein (GFP) reporter transgene to the domains of dlxexpression in the telencephalon and diencephalon (Fig. 6G,H). In this transgene construct, GFP is placed immediately downstream of a 3.5-kb fragment of the dlx6a 5′-flanking region including the promoter and part of the 5′UTR. This 5′-flanking fragment does not, by itself, target expression of GFP in a specific manner (Fig. 6F; no reproducible pattern in >150 embryos injected). However, in the presence of the zebrafish enhancers, 75–80% of injected embryos (n>400) had forebrain expression starting at 18 h postfertilization (hpf) and lasting until at least 96 hpf. Three transgenic lines could be produced all with comparable expression patterns and intensity. An embryo from one line is shown in Figure 6G and H. In contrast, the same intergenic fragment coupled to the ß-globin minimal promoter, which was used for transgenic mouse constructs, showed forebrain expression in only 8% of injected embryos and only 0.5% of them had more than 10 GFP-positive cells (Zerucha et al. 2000). The difference between efficiency of the human ß-globin minimal promoter fragment between human and zebrafish is, at present, unclear.
Similar transgene constructs containing the mouse I56i sequence (Fig.6I) or a combination of I56i and I56ii (Fig. 6J), inserted in the5‘-dlx6a-GFP plasmid, expressed GFP in the forebrain of transgenic zebrafish although the proportions of transgenic embryos were smaller than those observed with the corresponding construct containing zebrafish sequences. Thus, for both constructs, 35–40% embryos showed forebrain expression (n > 150 for each construct) with most of the GFP-positive cells in the telencephalic domain of dlx expression (Fig. 6 I,J).
The mouse I12b conserved sequence targeted reporter transgene expression to the forebrain of transgenic mice, starting at E10 and lasting until E16, the latest time point examined (Fig. 6C; Table 1; 3/3 primary embryos and 5/5 transgenic lines). This construct also produced expression in the apical ectodermal ridge, another site of endogenous Dlx expression although expression was more variable in intensity (Table 1) compared to that observed in the forebrain. Preliminary examination of sections of brains from lines of transgenic mice expressing the I12b-lacZ construct indicates that the constructs faithfully mimic expression of Dlx1/Dlx2in the telencephalon and diencephalon (data not shown). Thus, despite the fact that their sequences are highly divergent (see below), the three intergenic sequences, I56i, I56ii, and I12b, act ascis-acting forebrain enhancers with highly overlapping patterns of activity.
A 1.9-kb Xba1-EcoR1 fragment containing the I12a conserved sequence targeted lacZ expression to a subset ofDlx-expressing cells in the mesenchyme of the mandibular component of the first branchial arch and in the hyoid arch starting at E9.5 and lasting until at least E16, when expression gradually diminishes (Fig. 6D,E; Table 1; B.K. Park, S. Sperber, B.L. Thomas, G. Hatch, N. Ghanem, P.T. Sharpe, and M. Ekker, unpubl. observations). Reporter transgene expression was observed in six out of seven transgenic lines (Table 1). A 1.6-kb Xho1 fragment containing zebrafish I12a targeted expression in one out of two lines of transgenic mice (Table 1).
As the Dlx1/Dlx2 intergenic regions of mouse and human showed sequence conservation that extended beyond the above two enhancers (Fig. 3A), we produced transgenic mice with reporter constructs containing mouse intergenic fragments outside I12a and I12b. Thus, a construct containing a 1.5-kb DNA fragment located between I12a and I12b, with 80% identity between mouse and human (Figs. 1, 3A), did not show enhancer activity in mouse embryos (zero out of three primary transgenic embryos, as determined by detection of the transgene using PCR). Transgenic analysis of combinations of fragments from the mouseDlx1/Dlx2 intergenic region failed to indicate any enhancer activity that could be assessed to sequences outside I12a and I12b. Notably, some of these constructs included I12c (zero out of six PCR-positive embryos) suggesting that this sequence has no enhancer activity by itself, although it cannot be ruled out that it may cooperate with either I12a or I12b in a quantitative manner.
The Three Forebrain Enhancers Show Limited Sequence Similarity
The similar activity of the I12b, I56i, and I56ii enhancers in transgenic mice led us to investigate whether there could be sequence similarities between them. We made pairwise and dot matrix alignments of the three forebrain enhancers in both orientations. We also compared the forebrain enhancers with I12a. We did not find long stretches of sequence similarity among the four enhancers. The best dot matrix alignment was obtained by comparing I12b with I56i (Fig.7A). A short fragment that extended between 60–80 bp, depending on individual pairwise alignments, was present in all three forebrain enhancers but not in I12a. The two enhancers from the Dlx5/Dlx6 locus are in opposite orientations in this alignment (shown for the zebrafish sequences in Fig. 7B). The overall similarity over the short region is between 50–60%, thus smaller than the similarity between orthologous enhancer sequences (Figs. 1B, 2B). Interestingly, this region of similarity was also found downstream of the zebrafish dlx2b gene, a gene thought to be a duplicate ofdlx2a, but that is not part of a bigene cluster (A. Amores and M. Ekker, unpubl. observations).
Limited similarity between intergenic forebrain enhancer sequences. (A) Dot matrix comparison of the zebrafish I12b and I56i. The main two regions of sequence similarity are shown in B as multiple sequence alignments between I12b, I56i, and I56ii, and a sequence downstream of the zebrafish dlx2b. A three out of four consensus is shown. Putative Dlx binding sites, (A/C/G/) TAATT (G/A) (C/G), are indicated in bold, with mismatches highlighted. Additional TAAT/ATTA core homeodomain protein-binding sites are also highlighted.
The sequences shown in Figure 7B include a putative Dlxbinding site, (A/C/G/) TAATT (G/A) (C/G) (Feledy et al. 1999), near both ends of the similarity region. The core binding site for many homeodomain proteins (TAAT/ATTA) was also found between the two putative Dlx binding sites in many of the enhancers (Fig. 7B). The spacing between the Dlx binding sites was similar in all three enhancers. We previously showed that mutagenesis of both Dlx binding sites in I56i abolished almost completely the reporter gene expression in the forebrain of transgenic mice, suggesting that these sites are essential for activation or maintenance of enhancer activity, possibly through a crossregulatory or autoregulatory mechanism (Zerucha et al. 2000). The Dlx binding sites and surrounding nucleotides are less conserved in I56ii than those in I12b and I56i. The I56ii sequence is not activated by Dlx proteins in transfection assays, contrarily to I56i and I12b (Zerucha et al. 2000; N. Ghanem and M. Ekker, data not shown). This may also explain why it is less efficient than the other two enhancers in targeting a strong and consistent forebrain expression.
We also looked for additional protein-binding sites within the four enhancers (using Genomatix, Matinspector professional software;www.genomatix.de) and could not find any that were consistently found in all of them or in the three forebrain enhancers except for the homeodomain protein-binding sites TAAT/ATTA. Interestingly, theDlx binding site is also a low affinity-binding site (Chen and Schwartz 1995) for members of the Nkx family, that are known to be expressed in the forebrain. Nkx2.1, for instance, regulates regionalization in a subset of cells in the basal ganglia (Sussel et al. 1999) where the Dlx genes are also expressed.
In summary, the similarity between enhancers from paralogous bigene clusters occurs only in a small region of the total enhancer sequence, which, in turn, is highly conserved and over a much longer distance between orthologous, but not paralogous loci.
DISCUSSION
Conserved Organization of the Intergenic Region of Orthologous Dlx Bigene Clusters
We have performed a search for homologies in the intergenic region separating the two Dlx genes of bi-gene clusters in five different vertebrate species. Our analysis further illustrates the usefulness of “phylogenetic footprinting” (Muller et al. 2002) to identify cis-acting regulatory sequences. Examination of the region that separates the two Dlx genes that constitute theDlx1/Dlx2 or the Dlx5/Dlx6 bigene clusters reveals regions of high sequence conservation as well as conserved organization of the intergenic region for orthologous loci of distantly related vertebrates. Each of the two bigene clusters contains two regions of high sequence conservation that extend over a few hundred base pairs as well as a few shorter regions of sequence similarity. For both bi-gene clusters, the relative position and orientation of the conserved intergenic sequences are identical in all five species (Fig. 1, Fig. 2, and deposited sequence data).
The use of compact genomes found in tetraodontid species, such as the two pufferfish Takifugu rubripes and Spheroides nephelus was initiated to facilitate the search for regulatory elements. This is mainly because large regions of neutral DNA were lost in the course of genome reduction in these species, leaving the noncoding DNA regions enriched for cis-acting regulatory elements. We found that the presence of highly conserved sequences inDlx intergenic regions probably contributes to maintain its size even in species with compact genome. Thus, the size of theDlx1/Dlx2 and of the Dlx5/Dlx6 intergenic regions in the two pufferfish, although smaller than their mammalian counterparts, does not follow, proportionally, the smaller size of the genome of the two species.
Orthology assignment for the vertebrate Dlx genes was sometimes made difficult by the high degree of sequence similarity in the coding region of Dlx genes and by their highly overlapping patterns of expression. Conserved synteny, particularly with theHox clusters, was useful in establishing orthology relationship, as the Dlx bigene clusters have been found consistently on the same chromosome as one of the Hox clusters (Stock et al. 1996; Amores et al. 1998). Here, we propose that the sequence of the intergenic region is also a reliable predictor of orthology as the paralogous intergenic sequences are quite different while orthologous bigene clusters contain highly conserved sequences.
We examined whether or not the above prediction also applies to a duplicate gene in zebrafish: dlx2b (previously, dlx5; see comments about nomenclature in Methods). This gene shows high sequence similarity with members of the Dlx2 and Dlx5orthology groups. Mapping of dlx2b indicates that it is found in a group of genes with conserved synteny and that are a duplicate of a chromosome region that includes dlx2 (Amores et al. 1998). We examined about 8 kb of DNA downstream of dlx2b and found some sequence similarity with the noncoding sequence elements located in the Dlx1/Dlx2 intergenic region. Thus, sequences similar to I12a, I12b, and I12c were found (Fig. 7B and supplementary Figs. 6 and 8) although similarity was generally lower than when comparing individual elements between species. No sequence was found that resembled the conserved elements from the Dlx5/Dlx6 intergenic region except for the short sequence shown in Figure 7B. Thus, in addition to synteny analysis, conservation of noncoding sequence elements can be useful in establishing relationships between duplicate genes.
Highly Conserved cis-Acting Regulatory Sequences in the Intergenic Region of Dlx Bigene Clusters
The largest conserved sequences found in the Dlx1/Dlx2 andDlx5/Dlx6 intergenic regions are also the only ones conserved in all five species that were examined in the present study. The role of each of these sequences as a cis-acting regulatory element is demonstrated by their ability, once coupled to a promoter to drive expression of a reporter transgene in a tissue- and stage-specific manner. Sequence comparisons between mouse and human, or betweenTakifugu and Spheroides, reveals an overall high degree of sequence similarity and are therefore of less predictive value in the identification of regulatory elements. This may be because of the small evolutionary distance between the two mammals (∼50–60 Mya) as well as the two pufferfish (∼5–35 Mya), and to the slow rate of divergence for neutrally evolving regions among vertebrates in general (0.1% to 0.5% per million years) (Tautz 2000). Intergenic fragments outside the enhancers with 75–80% overall conservation between mouse and human failed to act, by themselves, as enhancers when tested in transgenic mice. Therefore, caution should be exerted when identifying putative cis-acting sequences based on comparisons between vertebrates of the same order. Comparisons that include multiple species with some that are distantly related might be a more efficient approach to identify noncoding sequence elements of functional importance, while keeping in mind that absence of sequence conservation does not necessarily indicate absence of functional conservation (Flint et al. 2001).
The relatively high degree of sequence conservation between the mouse and human Dlx1/Dlx2 intergenic region (80%) orDlx5/Dlx6 intergenic region (78%) contrasts with theDlx3/Dlx7 intergenic region that is only 69% identical, overall, between the two species (Sumiyama et al. 2002) despite the presence of sequences with higher percentage identity that may have a regulatory function (Sumiyama et al. 2002). However, comparisons of the mammalian Dlx3/Dlx7 intergenic region with those of zebrafish (Sumiyama et al. 2002), or Takifugu rubripes (N. Ghanem and M. Ekker, unpubl. observations) did not show conserved sequences comparable in length or percent identity to the four enhancers that we identified in the Dlx1/Dlx2 or in the Dlx5/Dlx6bigene clusters. Therefore, the Dlx3/Dlx7 bigene cluster may differ from its two paralogous Dlx clusters by a relatively low importance of the intergenic region in the mechanisms that control gene expression or by a higher divergence in regulation mechanisms between the different vertebrate lineages. Consistent with this latter hypothesis is the observation that zebrafishdlx3/dlx7 have marked differences in their early patterns of expression compared to their mammalian orthologs (Quint et al. 2000).
Function of Intergenic Elements in Dlx Regulation and Evolution
The organization of distal-less-related genes in bigene clusters may have preceded the evolution of vertebrates as two of the three characterized Dll genes of the ascidian Ciona intestinalis, Dll-A, and Dll-B are organized similarly with a short intergenic region (Di Gregorio et al. 1995). Recently, an enhancer located upstream of Dll-A was identified and shown to recapitulate most aspects of the endogenous expression pattern (Harafuji et al. 2002). Enhancers have yet to be found in the intergenic region that separates the Ciona Dll-A andDll-B genes and preliminary sequence comparisons did not reveal similarities in sequence between this region and the fourcis-acting regulatory sequence found in vertebrateDlx genes (M. Ekker, unpubl. observations).
Although the three Dlx bigene clusters of vertebrates are likely the result of duplication of an ancestral bigene cluster, we did not observe a high degree of conservation between paralogs, regardless of the species. This extends the observation previously made by Sumiyama and collaborators who compared the three human bigene clusters (Sumiyama et al. 2002). This lack of sequence similarity between paralogs is surprising, considering the similarities in expression patterns of genes found in paralogous bigene clusters.
Enhancers with overlapping patterns of activity (Fig. 6) show only a limited conservation in sequence (Fig. 7) that contrasts sharply with the high degree of conservation between orthologous sequences. Furthermore, enhancer sequences found in one Dlx bigene cluster are not found in the two paralogous clusters. Although one or several Dlx intergenic enhancers could originate from a sequence found in the ancestral Dlx bigene cluster, they would have diverged following the duplication events that took place early in vertebrate evolution, and that led to the three Dlx bigene clusters of modern vertebrates. This divergence happened before the separation of the lineages leading to modern-day teleost and tetrapods. Since then, purifying selection maintained most, if not all, regulatory mechanisms that involve these intergenic sequences, at least for theDlx1/Dlx2 and Dlx5/Dlx6 bigene clusters. The region of limited similarity found between the three forebrain enhancers may suggest that they resulted from a tandem duplication (I56i and I56ii) that also predated the split between the ray-finned fish lineages, and/or represent what subsists from a sequence present in the ancestralDlx bigene cluster.
Although the current study suggests that cis-acting regulatory elements of diverse sequence may exert similar enhancer function, the converse may also be true. Thus, I56i from mouse targets expression of a reporter transgene to the forebrain and mesenchymal cells of the branchial arches (Fig. 6A) whereas the orthologous sequence from zebrafish only directs expression to the forebrain, in either transgenic mice or zebrafish (Zerucha et al. 2000) despite the fact that the two sequences are >80% identical (Fig. 2B). Thus, the small differences in sequence between the enhancers from the two species may have a profound effect on enhancer function.
Evidence has been previously presented for cross-regulatory interactions between Dlx genes. Thus, the Dlx1 andDlx2 genes are expressed earlier in the forebrain and are involved in either the activation or maintenance of Dlx5 andDlx6 expression through the enhancer(s) found in theDlx5/Dlx6 intergenic region (Zerucha et al. 2000). In contrast, there is, at present, no evidence that Dlx5/6regulate Dlx1/2 in the brain. In the branchial arch mesenchyme, Dlx5/6 regulate Dlx3, but notDlx1/2 (Depew et al. 2002). Thus, the divergence of the intergenic enhancer sequences may have contributed to the specificity of cross-regulation between Dlx genes, allowing for sequential expression of paralogs.
The present study indicates an important role for the intergenic region in the cis regulatory mechanisms that are responsible for many aspects of the expression of genes from two Dlx bigene clusters. Intergenic regulatory elements are not solely responsible forDlx regulation. Thus, a fragment of the 5′-flanking region of mouse Dlx2 was shown to recapitulate expression in the epithelial cells of the branchial arches (Thomas et al. 2000). A targeted mutation, that inactivates the function of the mouseDlx1 and Dlx2, eliminates the entire intergenic region (Anderson et al. 1997). Intriguingly, homozygous mutants expressed truncated Dlx1 transcripts in the forebrain despite the absence of the I12b sequence (Zerucha et al. 2000). Although our results indicate that I12b is sufficient to confer expression of a reporter transgene to the forebrain (Fig. 6C), distinct sequences located upstream of Dlx1 also share this property (N. Ghanem and M. Ekker, unpubl. observations), suggesting a cooperative or synergistic effect between multiple and distinct enhancers in forebrain regulation of Dlx1 and/or Dlx2. Distinct mechanisms may take place at the Dlx5/Dlx6 locus. The lacZreporter gene, introduced in a targeted mutation of Dlx5/Dlx6that also removes the intergenic sequence (including I56i and I56ii), is only weakly expressed in the forebrain (Robledo et al. 2002). This suggests that enhancers outside the intergenic region may exist but that the intergenic enhancers play an essential role in conferring proper levels of gene expression, in as much as detection of transcripts by in situ hybridization can be considered quantitative. Taken together, these observations suggest complex mechanisms ofDlx expression control. These mechanisms involve multiple enhancers with overlapping but not necessarily redundant activity and a high degree of conservation in distant vertebrates for at least some of these enhancers.
METHODS
Dlx Gene Nomenclature
To help standardize the nomenclature for vertebrate Dlxgenes, we found it useful to adopt what was recently suggested byPanganiban and Rubenstein (2002). As the Dlx genes are found in regions of conserved synteny that contain the Hox clusters, the new nomenclature is aligned with that of the zebrafish hoxclusters (Amores et al. 1998). Thus, the zebrafish gene we refer to asdlx5a in this study is the gene previously named dlx4(Akimenko et al. 1994). Similarly, the zebrafish gene previously nameddlx5 is renamed dlx2b, as it is a dlx2duplicate (see Discussion). The previous dlx1, dlx2, and dlx6 genes are renamed dlx1a, dlx2a, anddlx6a, respectively. Finally, the previous dlx3,dlx7, and dlx8 genes of zebrafish would be renameddlx3b, dlx4b, and dlx4a, respectively. We kept the Dlx3/Dlx7 nomenclature for the mouse genes throughout the current report for the sake of simplicity but indicated the suggested name change.
Isolation and Characterization of Dlx Genes FromSpheroides Nephelus
Clones from a PAC library (Amemiya et al. 2001) were screened using a PCR approach for a conserved region of Dlx genes (Stock et al. 1996). The PCR fragments were sequenced to establish a preliminary orthology assignment. Genomic fragments comprising intron B and exon 3 of positive Dlx clones plus the intergenic region betweenDlx genes were obtained by PCR amplification using either specific or degenerate oligonucleotides.
Sequence Analysis
The zebrafish, mouse, and Spheroides intergenic sequences were determined from previously isolated genomic clones (McGuinness et al. 1996; Ellies et al. 1997; Depew et al. 1999) or from theSpheroides clones described in the above paragraph. They are deposited in GenBank under accession nos. AY168007–AY168012. The sequences from human and Takifugu rubripes were obtained from public databases: Human Dlx1/Dlx2, GenBank accession no. NT_005332.9; Human Dlx5/Dlx6, GenBank accession no. NT_033964.1; Takifugu dlx1/dlx2, scaffold 21, position 120318 to 125668, Takifugu dlx5/dlx6, scaffold 3932, position 6627–10192. For the Fugu Genome Consortium/JGI (DOE Joint Genome Institute), see http://www.jgi.doe.gov/index.html.
Pairwise sequence alignments are performed with PIPMAKER (available at http://bio.cse.psu.edu/pipmaker/), or with the BestFit, and Mapplot programs of the GCG Wisconsin package. Multiple sequence alignments are performed with the Pileup and Clustal X programs.
Transgenic Animals
For transgenic mice, sequences from the Dlx intergenic regions were subcloned into the p1229/p1230 vectors (Yee and Rigby 1993) that contain a human β-globin minimal promoter and thelacZ reporter gene. For transgenic zebrafish, intergenic enhancer sequences were inserted into a plasmid containing the GFP reporter gene placed downstream of a 3.5-kb fragment from the immediate 5′-flanking region of zebrafish dlx6a, including part of the 5′UTR. This fragment by itself, does not produce any tissue-specific expression in transgenic zebrafish (Fig. 6F). Subclonings were done using either a PCR-based approach or using convenient restriction sites. Transgenic animals were produced and analyzed as previously described (Zerucha et al. 2000).
WEB SITE REFERENCES
http://www.jgi.doe.gov/index.html; Department of Energy Joint Genome Institute. Genomic resources for Takifugu rubripes,Ciona intestinalis, and other species.
http://bio.cse.psu.edu/pipmaker/; Pipmaker computes alignments of similar regions in two DNA sequences.
www.genomatix.de; software and services including the MatInspector program to search for transcription factor binding sites.
Acknowledgments
We thank Luc Poitras and Fabien Avaron for useful discussions and Adrianna Gambarotta and Lucille Joly for technical assistance. N.G. was supported in part by a scholarship from the Lebanese University, Beyrouth. This work is supported by grants from the Canadian Institutes of Health Research (MOP14460) and the March of Dimes Birth Defects Foundation (FY01–207). M.E. is an Investigator of the CIHR.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
Present address: 5Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, TN 37232, USA; 6Department of Oral Anatomy, School of Dentistry, Chonbuk National University, Chonju, Republic of Korea.
-
↵7 These authors contributed equally to this work.
-
↵8 Corresponding author.
-
E-MAIL mekker{at}ohri.ca; FAX (613) 761-5036.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.716103.
-
- Received August 16, 2002.
- Accepted January 28, 2003.
- Cold Spring Harbor Laboratory Press