Lymphopenia in the BB Rat Model of Type 1 Diabetes is Due to a Mutation in a Novel Immune-Associated Nucleotide (Ian)-Related Gene
- Armand J. MacMurray1,4,
- Daniel H. Moralejo1,4,
- Anne E. Kwitek2,
- Elizabeth A. Rutledge1,
- Brian Van Yserloo1,
- Paul Gohlke1,
- Sara J. Speros1,
- Ben Snyder1,
- Jonathan Schaefer1,
- Sabine Bieg1,
- Jianjie Jiang1,
- Ruth A. Ettinger1,
- Jessica Fuller1,
- Terri L. Daniels1,
- Anna Pettersson1,
- Kimberly Orlebeke2,
- Bruce Birren3,
- Howard J. Jacob2,
- Eric S. Lander3, and
- Åke Lernmark1,5
Abstract
The BB (BioBreeding) rat is one of the best models of spontaneous autoimmune diabetes and is used to study non-MHC loci contributing to Type 1 diabetes. Type 1 diabetes in the diabetes-prone BB (BBDP) rat is polygenic, dependent upon mutations at several loci.Iddm1, on chromosome 4, is responsible for a lymphopenia (lyp) phenotype and is essential to diabetes. In this study, we report the positional cloning of theIddm1/lyp locus. We show that lymphopenia is due to a frameshift deletion in a novel member (Ian5) of the Immune-Associated Nucleotide (IAN)-related gene family, resulting in truncation of a significant portion of the protein. This mutation was absent in 37 other inbred rat strains that are nonlymphopenic and nondiabetic. The IAN gene family, lying within a tight cluster on rat chromosome 4, mouse chromosome 6, and human chromosome 7, is poorly characterized. Some members of the family have been shown to be expressed in mature T cells and switched on during thymic T-cell development, suggesting thatIan5 may be a key factor in T-cell development. The lymphopenia mutation may thus be useful not only to elucidate Type 1 diabetes, but also in the function of the Ian gene family as a whole.
[Sequence data reported in this paper has been deposited in GenBank and assigned the following accession nos:AF517674, AF517675, AF517676, and AF517677. Supplemental material is available online at http://depts.washington.edu/rhwlab/ and http:www.genome.org. ] The following individuals and institutions kindly provided reagents, samples, or unpublished information as indicated in the paper: K. Matsumoto and the Sir Frederick Banting Research Centre.
Type 1 (insulin dependent) diabetes mellitus in humans is a significant health problem with a prevalence ranging from 0.3% to 1% in different populations (Onkamo et al. 1999). Genetic studies in the human, mouse, and rat have shown that there are many genetic factors contributing to Type 1 diabetes besides the major histocompatibility complex (MHC) (Nerup et al. 1974; Todd et al. 1987;Graham et al. 2002). In the diabetes-prone BB rat (BBDP), we have identified three loci contributing to Type 1 diabetes outside the MHC complex on rat chromosome 20; Iddm1 on rat chromosome 4 (Jacob et al. 1992), Iddm3 on rat chromosome 2 (Klaff et al. 1999), and a fourth factor on rat chromosome 15 (Kwitek et al. 2002). TheIddm1/lyp locus, linked to peripheral T cell lymphopenia (<15% normal T-cell count) and Type 1 diabetes, was mapped to a 0.7-cM interval on rat chromosome 4 (Jacob et al. 1992), and the genetic mapping has been replicated many times (Hornum et al. 1995;Kloting and Kovacs 1998; Klaff et al. 1999). One notable feature about the BBDP diabetes model is that lymphopenia is essential for the development of the diabetic phenotype and is inherited as a simple Mendelian trait (Jacob et al. 1992; Bieg et al 1998). To study theIddm1/lyp locus in the absence of the other Iddmloci, we generated a congenic strain (DR.lyp), in which lymphopenia (lyp) and Iddm1 from a line of inbred diabetes-prone BB rats (BBDP) (Eastman et al. 1991; Markholst et al. 1993) was introgressed onto the genome of inbred diabetes-resistant BB rats (BBDR) by marker-assisted selection (Bieg et al. 1998). This congenic rat strain has confirmed that a single locus is responsible for both T-cell lymphopenia and spontaneous autoimmune diabetes. In the completed congenic DR.lyp line, and in recombinant animals developed from this strain, no animal developed diabetes without lymphopenia (lyp) (Bieg et al 1998). This suggests that either pleiotrophy is responsible for both traits or that the lymphopenia gene is responsible for the loss of critical T cells, resulting in autoimmunity. Specific pathogen-free wild-type (+/+) and heterozygous (lyp/+) DR.lyp rats have normal lymphocyte numbers and do not develop diabetes, whereas DR.lyp/lyp rats have T-cell lymphopenia from birth and clinical onset of Type 1 diabetes between 50 and 108 days of age in 100% of the animals (Bieg et al. 1998; Klaff et al. 1999). The nature of the Iddm1/lyp gene is therefore critical to the understanding of age-dependent Type 1 diabetes development in the BB rat.
In the BBDP and DR.lyp strains, Iddm1/lyp is a single Mendelian trait; therefore, we set out to identify it by using a positional cloning approach. We generated genetic and physical fine-structure maps of the region to identify and evaluate positional candidate genes for Iddm1/lyp. Here, we report the identification of a single nucleotide deletion in a novel member,Ian5, of the immune-associated nucleotide gene family, resulting in a largely truncated protein.
RESULTS
Comparative Genomics of the Iddm1 Genomic Interval
When we began the positional cloning of Iddm1/lyp, physical maps were not available for the rat. Therefore, we began by constructing a physical map of the syntenic region in the mouse, with the expectation that the mouse ortholog of Iddm1/lyp would lie within this interval. Comparative mapping determined that theIddm1/lyp region on rat chromosome 4 (between D4Mit6and D4Mit24) is syntenic to the proximal end of mouse chromosome 6 (Mmu6). Gene order appears to be conserved between rat and mouse, over the region just proximal of the ratIddm1/lyp region (including the T-cell receptor β-chain genes) to 10–15 cM distal of the locus (including the immunoglobulin κ chain complex, Igk) (data not shown). Because we expected the mouse Iddm1/lyp region to contain the ortholog of the ratIddm1/lyp gene, we set out to isolate the genomic DNA of both the mouse and rat Iddm1/lyp regions, combining the information and reagents from both species to create a comprehensive map of the region.
We initially constructed a mouse YAC contig spanning the ∼2-Mbp interval of the mouse Iddm1/lyp region, and isolated gene fragments from that interval. We then used the mouse gene fragments as probes to isolate the corresponding orthologous rat gene fragments by cross-species cDNA selection (Lovett et al. 1987). Next, we constructed a rat YAC contig spanning the rat Iddm1/lyp region by isolating those rat YAC clones that contained the rat gene fragments. STS content mapping and hybridization of gene fragments from one map to the other confirmed that the local gene order was the same in rat and mouse (these and other supplementary data are available on the authors' web sitehttp://depts.washington.edu/rhwlab/ and athttp://www.genome.org.).
Characterization of the Rat, Human, and Mouse lyp Regions
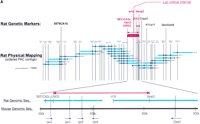
The initial mouse physical map was converted into a more useful higher-resolution form by isolating contigs of genomic BAC clones. We sequenced overlapping mouse BAC clones spanning >800 kb of the mouseIddm1/lyp region. STSs from these BAC clones were then used to refine the rat physical map by identifying corresponding rat PAC clones (rat contig shown in Fig.1a), which were then sequenced.
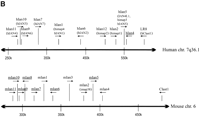
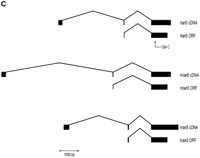
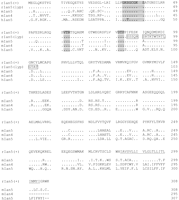
The lymphopenia gene region and the Ian5transcript. (A) Physical map of the rat lyp gene region with genetic markers integrated (top). Overlapping PAC clones are shown along with the locations of genetic markers used to narrow the lyp interval, clone-end STS assays, and the limits of the lyp interval itself as a red arrow at both topand bottom. Distances between markers may not be strictly to scale because they are estimated on STS content. The bottompart shows an expanded view of the lyp interval, showing the locations of known genes and the extent of the assembled sequence contigs of rat genomic DNA, along with the framework of mouse genomic DNA sequence. A 13-kb-long rat genomic sequence contig includes the ratIan5 gene. Position coordinates shown are those from mouse sequence supercontig Mm6_WIFeb01_100. (B) The cluster ofIan-related genes. In human, this gene family is present on chromosome 7q36.1. In the mouse, it is located on proximal chromosome 6. We have also indicated the position of the human and mouse orthologs of the LR8/Clast1 (mouse accession no. AB031386) gene as location aids, although this gene is not in the IAN family. We have indicated various alternative names associated with each gene, and provisionally named previously unnamed members of the family as follows (these are indicated by underlines). For those genes without a common name, we have chosen to continue using the Ian gene nomenclature. To avoid confusion, in this work we will refer to the genes in this family by using the name of the mouse ortholog and a prefix, h, m, or r, to specify which species is indicated (e.g., hIan2 for the human ortholog of mouse Ian2, otherwise known as himap1). Genes given the same Ian designation in different species have been determined to be orthologs of each other. Genes with different designations do not show enough similarity to be deemed orthologs, with the exception of hIan7, which is orthologous to both mIan7 and mIan3. Genes in species without a clear ortholog in the other have been given unique Ian numbers (e.g., hIan12). Positions shown are within the respective contigs (accession nos. NT_007704.8‖Hs7_7861 for human and supercontig Mm6_WIFeb01_100 for mouse). (C) Diagram of the rat, mouse, and human Ian5 gene transcripts, with exon structure shown to scale. Beneath each transcript diagram is a diagram of the extent of the major ORF (the Ian5-coding region). Continued on the following page.
While generating the physical map of the Iddm1/lyp regions in mouse and rat, we also generated recombinant animals to refine the position of Iddm1/lyp on the rat genetic map (Table1). We continued to intercross BBDR (+/+) and DR.lyp (lyp/lyp) rats, as well as to backcross and then intercross DR.lyp (lyp/lyp) and F344 rats. These crosses provided >300 additional animals in addition to the ∼870 animals already analyzed (Jacob et al. 1992), totaling over 2400 meioses. Resulting recombinant animals identified theIddm1/lyp interval, flanked by an SSLP, UW33, on the proximal end and a SNP, IIsnp3, on the distal end (Fig. 1a). This region corresponds to ∼100 kb on the mouse genome.
Recombinant Genotypes Specify the Boundaries of the lypCritical Interval in the Rat
With the recent assembly of a draft sequence of the mouse genome by the International Mouse Genome Sequencing Consortium (http://www.ensembl.org/Mus_musculus; Batzogloo et al. 2002), and other draft mouse contigs (e.g., MGSCv3 as available athttp://ncbi.nlm.gov/genome/seq/MmBlast.html) we integrated our sequence to produce a contig including the entire mouse region orthologous to the rat lyp interval (Fig. 1b). We then aligned the mouse genomic sequence to the human syntenic region on chromosome 7q36.1 and evaluated the conserved genes annotated in both species.
A notable feature of this region is the presence of a family of at least 10 putative GTP-binding protein genes found only in this region of the human and mouse genomes, the Immune Associated Nucleotide (IAN) gene family (Krucken et al. 1999; Poirier et al. 1999; Daheron et al. 2001; Cambot et al. 2002; Stamm et al. 2002). Interestingly, allIan gene family members are located in a 300-kb interval within 7q36.1 and a more compact 120-kb region in the mouse. This may be a consequence of genomic rearrangement in the human Iangene region relative to the mouse Ian gene region, because of the two species' evolutionary divergence, as the number ofIan genes differs in each (10 in human, 11 in mouse), and there are breaks in the gene order of the orthologs between the species (for example, hIan12 has no ortholog in the mouse, and mIan3 is one of two orthologs of hIan7). We examined the region in the mouse genome corresponding to the criticalIddm1/lyp interval in rat and found that three IAN family members lay within this critical region (Fig. 1a). WhereasIan2 was expressed in the spleen (Krucken et al. 1997),Ian4 was only expressed at low levels but was not detected in any other lymphoid tissue (Daheron et al. 2001). The third gene, designated Ian5, has not been reported previously in mouse or rat. Interestingly, rat Ian3 is differentially expressed in thymus and spleen when comparing tissue from DR.lyp+/+ andlyp/lyp rats (data not shown), but was excluded as a candidate because it lay outside of the critical region.
Identification of rIan5 that Contains a 1-bp Deletion Unique to the DR.lyp Rat
The intron/exon structure of the rIan5 gene is shown in Figure 1c in comparison with its mouse and human orthologs, mIan5 and IAN4L1 (hIan5). The overall genomic structure is similar to that reported previously in this family of genes (Stamm et al. 2002). As with hIan5, rIan5has at least three exons. The first and second exons are short, 220 and 49 bp, respectively, whereas the last exon is 1047 bp. There is a 3895-bp intron between exons 1 and 2, and a 1457-bp intron between exons 2 and 3. Exon 2 contains the putative start site for the major ORF spanning exons 2 and 3, as reported previously for mIan4(Daheron et al. 2001). Exons 1 and 2 contain an additional 61 amino acid ORF starting at position 78; this overlaps the major ORF and has no significant amino acid sequence similarity with the small 5′ ORF inmIan4.
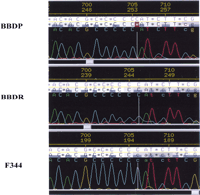
We established that rIan5 is expressed in rat spleen, thymus, and lymph nodes, making it a strong positional candidate forIddm1/lyp in the BB rat. To identify potential functional variants in this gene in the DR.lyp rats, we used primers spanning the putative coding sequence to amplify and sequence the gene from both BBDR wild-type and DR.lyp (lyp/lyp) thymic cDNA. The cDNA sequence was confirmed by sequencing of BBDP/WorAp and BBDR as well as F344 rat genomic DNA (Fig.2). The sequence analysis showed that both the DR.lyp and BBDP/WorAp strains lack one C nucleotide at basepair position 478 of rIan5, causing a frameshift mutation in the presumed ORF (exon 3) and leading to a significantly truncated predicted protein product (Fig. 3). The frameshift deletion in the lyp/lyp rIan5 changes the predicted downstream amino acids to include 19 amino acids (boxed) before the premature STOP codon (Fig. 3). We confirmed that this nucleotide deletion was present in our lymphopenic congenic F344.lyp inbred rat line (data not shown) as well as in outbred BBDP (diabetes prone) rats from Ottawa (Table2). As expected, the nonlymphopenic, diabetes-resistant outbred BBDR rat from Ottawa did not contain this deletion.
Sequence of the BB rat Immune Associated Nucleotide (Ian5) gene. A representative sequencing trace of DNA from BBDP/WorAp compared with wild-type BBDR/WorAp and F344 rats. The frameshift mutation at nucleotide position 478 in the DP rat DNA is indicated. DNA sequences were determined on an ABI PRISM 3700 DNA Analyzer (Applied Biosystems) and analyzed by the use of Phred, Phrap,Consed, and PolyPhred for sequence assembly and identification of sequence variants.
Sequence comparisons between BBDR wild-type (+/+), lyp/lyprat, mouse, and human Immune Associated Nucleotide-5 (Ian5) and mouse Ian4 predicted amino acid sequences. The deletion of a nucleotide in the codon for amino acid 85 of the rIan5(lyp) changes the predicted downstream amino acids to include 19 amino acids (italicized) before the premature STOP codon at amino acid position 104. Putative ATP/GTP-binding sites are boxed/shadowed and a hydrophobic putative transmembrane region underlined.
Sequence Analysis of Ian5 in Different Inbred Strains of Rats
To determine whether the frame shift deletion was a common polymorphism among rat strains or mutation unique to strains with lymphopenia, we resequenced ∼500 bp of rIan5, encompassing the deletion, in 38 rat strains (Table 2). The different strains have been characterized with genetic markers spanning the genome and were selected to represent inbred lines or strains of rats with maximum genetic diversity (Steen et al. 1999); only the BBDP/Ottawa and BBDP/WorAp strains have lymphopenia and Type 1 diabetes. The frameshift mutation was found only in the strains with lymphopenia (BBDP/Ottawa and BBDP). Three other sequence variants were found among the 38 strains and can be summarized as three distinct haplotypes (Table 2). The most common haplotype was found in 26 of the 38 strains; the frameshift mutation occurs on this haplotype. Whereas the normal rIan5 sequence predicts a protein of 35 kD, the deletion mutant would represent a dramatically truncated protein of 11 kD.
rIan5del Expression Reduced in Hematopoetic Cells
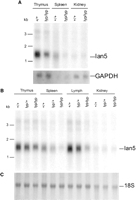
Northern blot analysis of poly(A+) RNA prepared from DR.+/+ and lyp/lyp rat tissues showed that the rIan5transcript of 1.4 kb was expressed in the thymus and spleen but not in the kidney (Fig. 4a). The transcript levels were markedly reduced in lyp/lyp as compared with +/+ tissue.Ian5 mRNA levels in thymus of lyp/lyp animals were reduced to 45% of wild-type levels. The level was even lower in the spleen of lyp/lyp animals (only 6% of wild-type levels), although this may reflect the absence of T cells resulting from the lymphopenia phenotype. Expression levels in the kidney were extremely low (3% of wild-type thymus), and differences between phenotypes could not be reliably observed. To prove that the decreased expression levels were a direct result of the frameshift mutation and not a secondary consequence of the absence of T cells due to lymphopenia, we examined expression levels in lyp/+ heterozygotes that show no lymphopenia. Heterozygotes showed intermediate levels of expression (Fig. 4b). These data support the notion that the frameshift mutation in the rIan5 gene causes a marked reduction in the mRNA in hematopoietic tissues established previously to be affected by T-cell lymphopenia and results in the lymphopenia and diabetes in the diabetic-prone BB rat. The reduced levels of rIan5 transcripts found in lyp/+ heterozygotes cannot be explained on the basis of T cell numbers, as both wild-type and heteozygotes have normal numbers of thymocytes and peripheral T cells, but may rather be due to the possibility that the mutated rIan5 transcripts are unstable.
Expression of rat Ian5 in tissues from lyp/lyp, lyp/+, and +/+ DR BB rats. (a) Northern blot containing 3 μg of poly(A+) RNA from thymus, spleen, or kidney of each of +/+ or lyp/lyp rats probed with a 695-bp region of Ian5showing a 1.4-kb transcript (Ian5). The blot was stripped and reprobed with a 1420-bp GAPDH probe (GAPDH). The images were quantitated using a PhosphorImager and software and normalized to GAPDH expression in each lane. Size markers are indicated. (B) Northern blot containing 3 μg of poly(A+) RNA from thymus, spleen, lymph node, and kidney from each of +/+, +/lyp, andlyp/lyp rats probed as in A. Size markers are indicated. (C) Methylene blue stain of the blot in Bbefore probing, showing even loading of 18S ribosomal RNA in each lane.
DISCUSSION
The BB rat has many important features as a model for the study of Type 1 diabetes (Crisá et al. 1992; Pettersson et al. 1996), including the presence of a simple mendalian trait, lymphopenia, which is absolutely essential for diabetes. In this work, we have shown that lymphopenia is due to a mutation in the novel rat Ian5 gene.
Ian5 belongs to a new and largely uncharacterized family of GTP-binding proteins that share sequence homology with the gene encoding the pathogen-induced plant protein aig1 (Reuber and Ausubel 1996). In Arabidopsis, this gene is induced after infection with Pseudomonas syringae. Another member of theIan family, IAP38, was induced in mice by blood-stage infections of Plasmodium chaubadi malaria (Krucken et al. 1997) and reported to be expressed in splenic macrophages, B cells, and T cells, providing some functional evidence that this family of genes plays a role in immune response.
The wild-type (DR) sequence of rIan5 predicts a protein with 308 amino acids, which is 80% identical to mIan5 and 52% identical to hIan5 (Daheron et al. 2001; Stamm et al. 2002). In the mouse, the coding regions of mIan5 and mIan4are 80% identical, although the noncoding portions of the genes are not substantially similar. In fact, amino acids 23–262 in mIan4 and amino acids 27–266 in mIan5 show only 7 amino acid differences (Fig. 3). The three species share an ATP/GTP-binding site at position 33–40 (Saraste et al. 1990). Other conserved regions for GDP/GTP exchange, GTP-induced conformational change, and GTP hydrolysis (Bourne et al. 1991), as suggested previously (Poirier et al. 1999; Cambot et al. 2002; Stamm et al. 2002), were identified at amino acids 61–63 and 81–83. A transmembrane domain at 282–304 was strongly predicted by TMHMM (Krogh et al. 2001).
The understanding of the genomic organization of this family of genes, as well as the function of the proteins, is as yet fragmentary as is the information about which cells express these proteins, and their subcellular localization. Apart from analyses in a variety of cell lines (Daheron et al. 2001), several of the Ian proteins are thought to be expressed in T or B cells, as well as in macrophages and dendritic cells. In contrast to our data on the rat Ian5 gene expression, mouse Ian4 was only expressed at low levels in the spleen and no expression was found in the thymus, liver, and kidney (Daheron et al. 2001). Mouse Ian5 has not been studied previously. Further studies are therefore required to identify the specific cell types in the BB rat that express the Ian5protein. The faint expression in the kidney may reflect Ian5expression in T cells.
The marked reduction in rIan5 expression in DR.lyp/lyp lymphoid organs may have several explanations. Lymphopenia in the BB rat is associated with a marked reduction in blood lymphocytes (Jackson et al. 1981; Bellgrau et al. 1982; Poussier et al. 1982; Elder and Maclaren 1983; Guttmann et al. 1983; Plamondon et al. 1990; Eastman et al. 1991) affecting both CD4+ and CD8+ T cells (Crisá et al. 1992), but in particular, T cells with the RT6 (ART2) T-cell marker (Greiner et al. 1997). Clearly, the reduction in T cells affects primarily peripheral blood, spleen, and lymph nodes, but not thymus (Gold and Bellgrau 1991; Bieg et al. 1997; Hernandez-Hoyos et al. 1999). The BBDP rat also exhibits abnormalities in the thymic T-cell subset (Groen et al. 1996), epithelial cell distribution (Doukas et al. 1994), and in vitro T-cell development (Whalen et al. 1999). The possibility that Ian5 is expressed chiefly in T cells is supported by the drastic reduction in expression in DR.lyp/lyp lymphoid organs. In this case, the observation that heterozygous DR.lyp/+ rats had levels ofIan5 that were intermediate between lyp/lyp and +/+, but had normal T-cell levels, is best explained by the possibility that the mutated rIan5 transcript is unstable.
It will now be possible to determine whether the truncatedIan5 is causing lymphopenia because of the loss of a transmembrane domain or because of a greater functional disruption to the protein. Future studies can also test the hypothesis thatIan5 may be a key factor in T-cell development. It will also be possible to begin the analysis of pathways downstream ofIan5 to uncover important checkpoints in the emergence of age-dependent autoimmune diabetes. All of this will greatly enhance the utility of the BB rat in dissecting mechanisms that control autoimmunity.
Finally, it will now be feasible to test whether this gene plays a role in human Type 1 diabetes by identifying SNPs in the gene and conducting association studies. Independent of the role this gene plays in human diabetes, its identification will increase our understanding of both the disease process and the role of non-MHC loci in diabetes.
METHODS
Rats
BBDR (Bieg et al. 1998) and F344 rats (Klaff et al. 1999) congenic for lymphopenia were maintained at the University of Washington. All animals were kept under specific pathogen free (SPF) conditions with standard light-dark cycles. The rats were fed a regular diet. Sentinel animals were negative for viral antibodies and parasites during the period of the experiments. Siblings heterozygous for polymorphic markers flanking the lymphopenia interval were used as breeding pairs to generate homozygous animals. The rats were screened for diabetes and lymphopenia as described in detail (Bieg et al. 1998).
DNA was obtained from 32 different rat strains as described previously (Kwitek et al. 2001). In addition, we analyzed DNA from LEA, LEC, OLETF, and WKAH rats kindly donated by Dr. Kozo Matsumoto and from outbred BBDR and BBDP rats from Health Products & Food Branch, Sir Frederick Banting Research Centre, .
Physical Mapping and STS Screening
Mouse YAC Contig
Mouse YAC contigs were generated by first screening with known STSs and then filling in gaps by sequencing YAC ends and using resulting nonrepetitive sequence as additional STSs. Unless pre-existing or otherwise noted, PCR primers were selected using the Primer 0.5 program (Lincoln et al. 1991) to choose primers with predicted melting temperatures within 1°C of 60°C and to avoid regions with repeat- or self-similarity. PCR amplification was performed according to the conditions specified for each protocol, or as described previously, or, if not specified, according to standard conditions as recommended by Perkin-Elmer.
YACs were isolated from the MIT mouse YAC library (Kusumi et al. 1993) by use of standard PCR screening methods and the YAC DNA prepared as described (Segre et al. 1995). YAC ends were isolated by use of inverse PCR as described previously (Haldi et al. 1994) and sequenced directly by use of standard fluorescent sequencing methods.
Higher Resolution Physical Mapping
Mouse bacteriophage P1 clones were isolated from two libraries, the P1 mouse RIII (2–3× coverage) and the P1 mouse ES (3× coverage) libraries (Pierce et al. 1992; Sternberg et al. 1994) (Genome Systems). Mouse BAC clones were isolated from a 129/SV mouse BAC library CITB CJ7B prepared by Bruce Birren (7× coverage) (Shizuya et al. 1992; Kim et al. 1996). Rat PAC clones were isolated from the RPCI-31 library (Woon et al. 1998). Each library was screened by a PCR-based or hybrid PCR- and hybridization-based protocol, as recommended by the library maker. P1, BAC, and PAC DNA was prepared according to standard protocols and as recommended by Genome Systems. P1, BAC, and PAC end sequences were obtained by use of a protocol similar to that for cloning YAC ends. STS content maps were assembled by use of standard PCR techniques to determine the STS content of panels of miniprep DNA from the isolated P1s, BACs, and PACs. Small rat genomic contigs were assembled by sequencing subclones, by directly sequencing PCR products, and by assembling contigs from the publicly-available rat genomic raw trace (WGS) database maintained by NCBI (http://ncbi.nlm.nih.gov/genome/seq/RnBlast.html).
Cross-Species cDNA Selection
Cross-Species cDNA Selection was performed by use of a protocol modified from that described previously by Lovett (1987, 1991) and the primers cDNA-1 (5′-CTGAGCGGAAT TCGTGAGACC-3′)/cDNA-2 (5′-P-GGTCTCACGAATTCCGC TCAGTT-3′). All mouse template cDNAs were separately PCR amplified 10–15 cycles (94°C/64°C/72°C) with the bio-cDNA-1 primer (5′-biotin-CTGAGCGGAATTCGTGAGACC-3′; 64.4°C predicted melting temperature) and purified. Double-stranded rat cDNA from testis and spleen with an average fragment size of ∼500 bp was modified with linkers composed of the two oligos cDNA-1b (5′-GTCACGCAAGCTTCT CACAGG-3′) and cDNA-2b (5′-P-CCTGTGAGAAGCTTGCGT GACTT-3′) and amplified using the cDNA-1b primer. A total of 1 μg-amplified cDNA, 2 μg mouse C0t-1 DNA (BRL), and 2 μg glycogen (BMB) were prehybridized to a C0t value four times greater than in the standard protocol. The prehybridized rat cDNA mixture was then mixed with the mouse template cDNA and hybridized essentially as in the standard protocol. After hybridization was stopped, the biotinylated material was washed 3 × 15 min in 0.1 × SSC/0.1% SDS at one of the three wash temperatures 65, 55, or 50°C (depending on the stringency desired). Finally, the selected cDNAs were eluted and eventually dU-cloned into the pAMP10 vector (BRL) by amplifying 30 cycles with 60°C annealing using the cDNA-U-2 primer (5′-CUA CUACUACUA GTCACGCAAGCTT CTCACAG-3′).
Genotyping
DNA was extracted from rat tail biopsies obtained at 25–30 days of age. Genotyping for simple sequence repeat markers was carried out as described previously (Jacob et al. 1992). Rat tail DNA was PCR amplified using IRD-700 tailed primers (LI-COR Biosciences). The PCR products were analyzed using a NEN Global IR2 DNA Analyzer System (Model 4200S-2) using 6.5% gel matrix (LI-COR Biosciences).
DNA Sequence Analysis
Initial Sequence Analysis of BB DR.lyp
Primer pairs were designed for amplification of therIan5-coding exons 2 and 3 ( forward primer, 5′-GCTTGAGGAGGTCATCAGTTC-3′ and reverse primer, 5′-CTCACGTC CCAGCCTCTAAC-3′). PCR reactions were 2 min at 95°C; 10×; 30 s at 95°C, 30 s at 60°C, 30 s at 72°C; 30× 30 s at 95°C, 30 s at 60°C, 30 s plus 10 s/cycle at 72°C; 7 min at 72°C. The PCR products were purified with Ultrafree-MC (Millipore). We subjected purified PCR products (30–60 ng) to cycle sequence reactions using IRDye 800 terminators (LI-COR) and Thermo Sequenase (USB). The reaction products were purified with a MultiScreen Filtration System (Millipore) using Sephadex TM G-50 Fine (Amersham Pharmacia Biotech) and analyzed using NEN Global IR2 DNA Analyzer System sequencer (LI-COR Inc.)
Sequencing of Additional Inbred Rat Strains
Primers were selected for PCR amplification of 500 bp (forward primer 5′-CCATGGCTTTGAGGAACTATCC-3′ and reverse primer 5′-TGTGGGTGAAGAGGACAATCAT-3′) and 385 bp (forward primer 5′-AAAGTGCCACAGGGAACAGC-3′ and reverse primer 5′-GTGTGGGTCACAAACTCTTCCA-3′) fragments, encompassing therIan5 deletion mutation. Amplified products were subject to standard fluorescent sequencing using an ABI3700 automated sequencer. Analysis was performed using Phred, Phrap,Consed, and Polyphred to compare the sequences between BBDP and 38 other inbred rat strains.
In Silico Sequence Analysis
For the human, we used NCBI's genomic TBLASTN with the predicted protein product of hIan5 blasted against the GenBank human genome as of 12/24/01, setting the expectation parameter to 10 (http://ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs). The E values of the resulting matches were bimodal, with the matches plotted in Figure 1b ranging from ê-167 to 4ê-28 and the remaining spurious matches having E values >1.
For the mouse, we again used TBLASTN with the predicted protein product of mIan4 blasted against the GenBank mouse genome supercontig database (mgscv3) posted on 4/19/02, setting the expectation parameter to 10 (http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html). Again, the resulting E values were bimodal, with the Figure 1b matches ranging from ê-160 to 3ê-8 and the remaining spurious match having an E value of 1.5. We also used other NCBI Blast programs such as BlastN and Blast2 according to recommended settings, to identify homologous EST sequences, already-identified genes, and the alignments of one sequence within another (Altschul et al. 1990, 1997);http://www.ncbi.nlm.nih.gov/BLAST/.
RNA Isolation and Quantification of Ian5 mRNA Levels
Organs were dissected from 49-day-old congenic DR.lyp rats of each of three genotypes (wild-type or +/+, lyp/+, andlyp/lyp) from the same litter and homogenized immediately. Poly(A+) RNA was isolated using QIAGEN RNeasy and Oligotex kits (QIAGEN). A total of 3 μg of poly(A+) RNA per well was electrophoresed through a 0.9% agarose gel containing MOPS buffer [40 mM 3-N-morpholino-propane sulfonic acid (MOPS), 10 mM sodium acetate, 1 mM EDTA), and 2% formaldehyde. The gel was washed twice, 30 min in DEPC water, 35 min in 50 mM NaOH, 1.5 M NaCl, 30 min in 1 M Tris (pH 8.0), 1.5 M NaCl, and 5 min in 10× SSC (1× SSC is 0.15 M NaCl, 15 mM Na citrate at pH 7.0). The RNA was transferred to a positively charged nylon membrane (Roche) by vacuum blotting and cross-linked to the membrane in a UV Stratalinker 1800 (Stratagene). The membrane was stained with methlyene blue stain (0.03% methylene blue in 0.3 M sodium acetate at pH 5.2).
Blots were prehybridized for 1 h in Church buffer (0.5 M Na phosphate buffer at pH 6.8, 1 mM EDTA, 7% SDS) at 65°C. 32P-labeled probe was made by amplifying a 695-bp fragment by PCR using rIAN4–690f (5′-CTCCTGGTGGGTAAATCTGG-3′) forward primer and rIAN4–1384r (5′-TCCTTCAGCTCCCT CTTCTG-3′) reverse primer (Invitrogen) in mix containing: 50 ng of genomic DR.lyp DNA, 1× TAQ Polymerase Buffer, 250 μM each (dATP, dGTP, dTTP) and 50 μM dCTP, 0.5 μM each primer, 40 μCi 32P-labeled dCTP (PerkinElmer Life Sciences), 0.5 U TAQ 2000 DNA Polymerase (Stratagene). Probe was amplified at 95°C for 3:00 min, then 35 cycles of 95°C for 0:45, 60°C for 0:45, 72°C for 1:00 min.
The probe was purified through a G50 AutoSeq column (Amersham Pharmacia Biotech), denatured by heating at 96°C for 7.5 min, iced, and added to the blot overnight at 65°C. The blot was rinsed twice with 2× SSC/0.1% SDS at room temperature, then washed with 2× SSC/0.1% SDS at 65°C for 20 min, 0.2× SSC/0.1% SDS at 65°C for 20 min, 0.1× SSC/0.1%SDS at 65°C for 30 min. The blot was then placed on BioMax MS Film (Eastman Kodak) and subsequently on a phosphorscreen to be scanned by a STORM 840 PhosphorImager and quantified withImageQuant v1.2 software (Molecular Dynamics).
Blots were stripped by an overnight wash in 0× SSC/0.1% SDS at 65°C and reprobed with rat GAPDH (Accession no. AB017801) cloned into pGEM3z. Using the same method described above, T7 (5′-TAATACGACTCACTATAGGG-3′) forward primer and T3 (5′-ATTAACCCTCACTAAAGGGA-3′) reverse primer were used to generate a 1420-bp fragment in mix containing 1 ng of pGEM3z-rGAPDH, 1× Taq Polymerase Buffer, 250 μM each (dATP, dGTP, dTTP), and 50 μM dCTP, 0.5 μM each primer, 40μCi 32P-labeled dCTP (PerkinElmer Life Sciences), 0.5 U TAQ2000 DNA Polymerase (Stratagene). Probe was amplified at 95°C for 3:00, then 35 cycles of 95°C for 0:45, 50°C for 0:45, 72°C for 1:45.
WEB SITE REFERENCES
http://depts.washington.edu/rhwlab/; Authors' web site for data from this paper.
http://rgd.mcw.edu; Rat genome database.
http://www.ensembl.org/Mus_musculus; International Mouse Genome Sequencing Consortium's assembly of the mouse genome.
http://www.ncbi.nlm.nih.gov/BLAST/; NCBI Blast home page.
http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html; NCBI Mouse Genome Blast web site, including MGSCv3 contig database.
http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsBlast.html&&ORG=Hs; NCBI Human Genome Blast web site, including latest human genome assembly.
http://www.ncbi.nlm.nih.gov/genome/seq/RnBlast.html; NCBI Rat Genome Blast web site, including WGS raw trace database.
http://www-genome.wi.mit.edu/; Whitehead Institute/MIT Center for Genome Research home page.
Acknowledgments
This work was supported by the National Institutes of Health (AI42380, Cardiopathology Training Grant T32-HL07312), the Molecular Genetics Core of the University of Washington Diabetes Endocrinology Research Center (DK17047), the Juvenile Diabetes Research Foundation (grant 1-2002-240), and a Junior Faculty Award (to R.A.E., grant 1-02-JF-06), as well as a Mentor Based Postdoctoral Fellowship Award (to Å.L.) from the American Diabetes Association. DNA from LEA, LEC, OLETF, and WKAH rats were kindly donated by Dr. Kozo Matsumoto, Institute for Animal Experimentation, The University of Tokushima, Japan. We thank Philippe Poussier and Hemmo Drexhage for comments to the manuscript and Michael Jensen-Seaman, Jeff Nie, and Paul Havlak for validation of sequencing data as well as Sue Blaylock for outstanding assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.