Vestibular Dysfunction in the Epistatic circler Mouse Is Caused by Phenotypic Interaction of One Recessive Gene and Three Modifier Genes
- Kim Cryns1,
- Michiel P. Van Spaendonck2,3,
- Kris Flothmann1,
- Arjan M. van Alphen4,
- Paul H. Van De Heyning3,
- Jean-Pierre Timmermans2,
- Chris I. De Zeeuw4, and
- Guy Van Camp1,5
- 1Department of Medical Genetics, University of Antwerp, B-2610 Antwerp, Belgium; 2Laboratory of Cell Biology and Histology, University of Antwerp, B-2020 Antwerp, Belgium; 3Department of Otolaryngology, University of Antwerp, B-2610 Antwerp, Belgium; 4Department of Anatomy, Neuroscience Institute, Erasmus University Rotterdam, 3000 DR Rotterdam, The Netherlands
Abstract
Vestibular dysfunction is a frequent clinical problem, leading to dizziness and imbalance. Genes play an important role in its etiology, but the genetics are complex and poorly understood. In this study we have analyzed the complex inheritance pattern in the Epistatic circler mouse, which shows circling behavior indicative of vestibular dysfunction in the mouse. This phenotype exists in a proportion of the F2-generation from an intercross betweenC57L/J and SWR/J mouse strains. Genetic investigation indicates that the circling behavior is caused by a major recessively inherited gene derived from the SWR/J strain (theEcs-gene) in combination with at least three different modifier genes derived from C57L/J (the Ecl-genes). Genetic mapping made it possible to localize the Ecs-gene to chromosome 14 and the Ecl-genes to chromosome 3, 4, and 13. This study illustrates the feasibility of identifying genes for multifactorial traits in mice.
The vestibular system plays a key role in postural reflexes (reflexes that stabilize the body such as forelimb extension, hindlimb flexion, and activation of head muscles) and in compensating eye movements of head displacement by means of the vestibulo-ocular reflex. This reflex is a mechanism that produces eye movements that counter head movements to stabilize the gaze. If the system is damaged, gaze stabilization, balance, and orientation in space are all affected. These manifestations of vestibular damage can have severe consequences in man such as vertigo, oscillopsia, and imbalance. Although vestibular dysfunction is seen in a few rare monogenic syndromes, it is commonly encountered in isolated form in otological practice, the etiology often remaining obscure. The importance of genetic factors in peripheral vestibular dysfunction has been stressed on numerous occasions (Verhagen et al. 1987; Baloh et al.1994), but little is known about the underlying genes. The most important reason why genetic studies of human genetic vestibular defects are difficult to perform lies in the lack of large monogenic pedigrees containing a sufficient number of affected family members to perform genetic linkage analysis. Therefore, vestibular dysfunction is thought to be a multifactorial condition involving interactions between multiple genes in most cases. The use of animal models, and in particular mouse models, is an alternative to genetic studies in human to gain more insight into the complex phenomenon of vestibular dysfunction.
In this study we have used the Epistatic circler mouse as a model for vestibular dysfunction. Doolittle (1963) was the first to describe the Epistatic circler mouse in the early 1960s. He reported that a small proportion of the F2-generation of the cross between C57L/J and SWR/J inbred mouse strains shows shaker-waltzer behavior. Shaker-waltzer behavior is a common phenomenon among mouse mutants and consists of hyperactivity, head shaking, and/or circling. It is caused by vestibular dysfunction and is often associated with structural anomalies of the vestibular apparatus. According to Doolittle (1963), the Epistatic circler mice circled either to the left or to the right, showed no head tremor, did not appear to be deaf, and swam well. He indicated that the circling behavior results from the X-yy genotype in which X is the dominantEcl-gene, derived from C57L/J, and y the recessiveEcs-gene, derived from SWR/J (Doolittle 1963). Alternatively, Taylor (1976) suggested that the circling behavior ofEpistatic circler mice is caused by the simultaneous homozygosity for two recessive genes. After outcrossing AKXLrecombinant inbred lines to SWR/J, Taylor found that the circling behavior in the F2-progenies showed concordant inheritance with the hexose-6-phosphate dehydrogenase locus on chromosome 4 in eight recombinant inbred lines, suggesting that the Ecl-gene is localized on chromosome 4. However, because Taylor used a different strain combination than Doolittle, it cannot be excluded that the different models are attributable to the strain combinations used. No detailed genetic investigation of the Epistatic circler mouse has been published before. In the present study we used the model proposed by Taylor as a starting point to perform mapping studies. Genetic mapping revealed that the circling behavior is caused by a combination of a major recessive Ecs-gene and three modifyingEcl-genes.
RESULTS
Intercross and Backcross
C57L/J mice were mated with SWR/J mice and an intercross was set up between the F1-progeny. Recovered were 3662 F2-progeny, which were scored phenotypically for their circling behavior at the age of 3 wk. Actively circling animals were scored as affected, and small pieces of their tails were cut off for DNA preparation. Of the 3662 mice, 115 were scored as affected, which corresponds to 1 in 32. Affected as well as unaffected F2-animals were fertile. Affected mice also showed hyperactivity without head bobbing. To verify that the circler mutants had normal hearing, we tested their Preyer reflex. Sound stimuli evoked a flick of the pinnae in all 115 circling F2-animals. In addition, hearing in mice was assessed by auditory brainstem response (ABR) measurements in two F2-circlers and two noncircling littermates, all at the age of 2 mo. Circling as well as noncircling F2-animals showed the same ABR pattern in response to a broad frequency range stimulus.
An additional backcross was set up between affected F2-animals and mice from the SWR/J strain. Recovered were 411 N2-progeny, which were tested for circling behavior. Eighteen animals showed circling behavior (1 in 23).
Homozygosity Mapping
Pooling DNA samples is a very efficient approach for mapping recessive genes and has been used previously in mice for mapping Mendelian and complex traits (Taylor and Rowe 1989; Asada et al. 1994;Alfred et al. 1997). It basically consists of reducing the number of PCR reactions and subsequent gel runs through analysis of a combined DNA sample obtained by pooling together equal amounts of DNA from affected animals. Based on segregation analysis, Taylor (1976)suggested that the circling behavior is attributable to two recessive genes. We used this model as a starting point for linkage analysis and used a DNA-pooling strategy to rapidly identify the chromosomal regions that carry recessive Ecl- and/or Ecs-genes. First, we analyzed which markers from the Mouse MAPPAIRS Genome-Wide Screening Set were polymorphic between C57L/J and SWR/J mice. Of 340 tested markers, 240 showed a different allele for both parental strains. These 240 microsatellite markers were dispersed over the entire autosomal portion of the genome and were analyzed on a combined DNA sample that was obtained by pooling equal amounts of DNA from 17 F2-circlers. To be sure that the distance between two markers was not >20 cM, eight additional markers were chosen from the Whitehead/MIT map. Of all 248 markers tested in the genome scan, two markers (D14Mit 120 and D14Mit 141) showed only the allele ofSWR/J in the affected DNA pool. Both markers were located in the same chromosomal region, suggesting that the Ecs-gene was located on chromosome 14. To confirm this result, we analyzed these two markers on the 17 separate DNA samples. All 17 F2-circlers were homozygous for the SWR/J allele of this microsatellite marker. χ-square testing gave a P-value <10−11 for both markers, in which a pointwise P-value <10−4 has been suggested as significant linkage for genome-wide studies in the mouse (expected to occur 0.05 times by chance in a genome scan) (Lander and Kruglyak 1995). LOD score calculations for both markers gave identical maximum LOD scores of 10.2 in which a LOD score of 3.3 has been suggested as a genome-wide significance limit (Lander and Kruglyak 1995). D14Mit44, a marker proximal toD14Mit120, and D14Mit32, a marker distal toD14Mit141, showed both alleles from C57L/J andSWR/J in the DNA pool. These results place theEcs-locus in a 24 cM region on chromosome 14 (based on the Whitehead/MIT map). None of the 248 informative markers showed only theC57L/J allele in the analysis of the DNA pool. Therefore, no markers linked to the Ecl-locus were detected, indicating that there is no recessive Ecl-gene.
Mapping of the Ecl-Gene
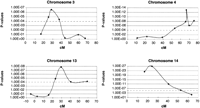
As homozygosity mapping did not yield any markers linked to theEcl-locus, we hypothesized that the Ecl-gene was a nonrecessive gene. To confirm this hypothesis, we set up an additional cross between F2-circlers and SWR/J mice. The offspring of this cross (N2) will never show homozygosity for the mutantEcl-gene, because it is derived from C57L/J. In consequence, if there are any circling animals in the N2-offspring of this cross, they will invariably show heterozygosity for the mutantEcl-gene, excluding the possibility of recessiveEcl-genes. Eighteen out of 411 N2-progeny showed circling behavior, confirming the nonrecessive character of theEcl-gene. To localize this gene, we separately genotyped 46 F2-circlers using 87 polymorphic markers dispersed over the entire autosomal portion of the genome. Again we included the same eight additional markers as in the homozygosity mapping, which were located in the regions with the largest intermarker distances. P-values were calculated for each marker. Of all markers tested, 22 markers showed a P-value of 0.05 or less. Five of these markers were localized on chromosome 14, confirming the linkage to the Ecs-locus previously found by means of the DNA-pooling strategy. With the exception of the chromosome 14 markers, the other 17 markers with a suggestive P-value were analyzed on 43 additional samples. Only seven markers showed a P-value of 10−4 or less (based on 89 samples), which indicates significant linkage (Lander and Kruglyak 1995). These markers were located on chromosome 3, 4, and 13. The most frequent allele was in each case derived from the C57L/Jstrain. For the localization on chromosome 3, 4, and 13, two nearby markers were analyzed on 89 samples to confirm linkage. All additional markers showed a P-value of 10−4 or less. Figure1 gives a graphical overview of the P-values from chromosomes 3, 4, 13, and 14.
Graphical overview of the P-values from chromosome 3, 4, 13, and 14. Centimorgans are plotted on the X-axis; P-values are plotted on the Y-axis. P-values ≤10−4 were considered as significant linkage for a genome-wide search (Lander and Kruglyak 1995). D3Mit224, D3Mit22, D3Mit230,D3Mit142, D3Mit217, D3Mit147, andD3Mit19 are plotted for chromosome 3. D4Mit171, D4Mit214, D4Mit166, D4Mit37, D4Mit148, D4Mit251, D4Mit170, andD4Mit63 are plotted for chromosome 4. D13Mit16, D13Mit207, D13Mit221, D13Mit186, D13Mit191, D13Mit213, and D13Mit35 are plotted for chromosome 13. D14Mit44, D14Mit141, D14Mit32, D14Mit194, and D14Mit75 are plotted for chromosome 14.
Liability Scale
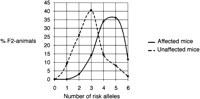
Because the Ecs-gene is a major determinant, necessary but not sufficient to cause circling, we wanted to investigate the contribution of the Ecl-genes by defining a liability scale to determine a threshold. To do this, we first genotyped 243 F2-noncirclers for the recessive Ecs-gene. Fifty of these unaffected animals inherited both copies of the mutantEcs-gene. In a next stage, we genotyped 47 of these selected animals for a genetic marker close to each of the threeEcl-genes (on chromosome 3, 4, and 13), and we counted the number of risk Ecl-alleles (the allele originating fromC57L/J). For genotyping, we chose markers with the most significant P-value (D3Mit230, D4Mit251, andD13Mit191). Table 1 gives an overview of the distribution of different genotypes at the threeEcl-loci. Note that animals that show two Ecla-risk alleles (chromosome 3) and one Eclc-risk allele (chromosome 13) but no Eclb-risk allele (chromosome 4), are unaffected (AAbbCc genotype). This genotype was never found in affected animals, whereas 8.5% of the unaffected animals displayed this genotype. Only one affected animal did not inherit an Eclb-risk allele (chromosome 4). Figure 2 gives a graphical overview of these results for a model of equal and additive contribution of the three Ecl-genes. Although there is a shift in the number of risk alleles between unaffected and affected F2-animals, there is no complete separation between affected and unaffected animals, and a clear threshold cannot be determined.
Distribution of the Different Genotypes at the ThreeEcl-loci in 91 Circling and 47 Noncircling Mice
Liability scale for equal and additive effects for theEcl-genes. The number of risk Ecl-alleles is plotted on the X-axis; the fraction of F2-animals (47 noncircling and 91 circling F2-mice) is plotted on the Y-axis. Although we were not able to determine a threshold, there is a clear shift in the number of affected alleles between unaffected and affected F2 animals.
DISCUSSION
In this paper we report on the genetic characteristics of theEpistatic circler mouse, an existing mutant with a vestibular pathology. The most overt behavior consists of rapid bidirectional circling and hyperactivity, which is indicative of vestibular dysfunction (Steel 1991). A positive Preyer reflex and normal ABR test results indicate that the hearing of the Epistatic circlermouse is not severely affected.
To map the responsible genes, we first performed a genome scan by means of a DNA-pooling strategy. This study revealed a single major recessive gene, the Ecs-gene, located on chromosome 14. An additional cross between F2-circlers and SWR/J mice indicated that theEcl-gene shows a nonrecessive character. Surprisingly, a genome search revealed that the circling behavior in the Epistatic circler mouse is under the control of more than two genes. Next to the recessive Ecs-gene, we found three modifyingEcl-genes localized at chromosomes 3, 4, and 13. We propose to call these genes Ecla, Eclb, and Eclc, respectively. We compared genotypes of circling and noncircling mice to define a model explaining the interaction between the different genes. Two important facts were noted. First, we never found circling mice that were not homozygous for the Ecs-gene, indicating that two risk alleles for Ecs are needed for circling. Second, we could identify only one affected animal that showed homozygosity for the nonrisk allele of the Eclb-gene. As the exact genetic distance between the marker used for genotyping and the Eclb-gene is unknown, a recombination between these two may have occurred. In this case, all affected animals have at least one risk allele forEclb, and one copy is probably obligatory for circling. The fact that Ecla and Eclc are clearly not obligatory suggests an epistatic interaction between Ecs (recessive) andEclb (nonrecessive), with additive effects ofEcla, Eclc, and possibly the second allele ofEclb. A simple model of equal and additive effects forEcl-genes is represented in Figure 2. The fact that we were not able to determine a clear threshold in the number of riskEcl-alleles indicates that this model is not (entirely) correct. Functional inequality between the three Ecl-genes may be an explanation, and the involvement of additional factors (genetic as well as environmental) also cannot be excluded. In conclusion, although other genes and environmental factors may be involved, it is clear that the Ecs-, Ecla-, Eclb-, andEclc-genes are major factors that cause vestibular dysfunction in the Epistatic circler mouse.
We mapped the Eclb-gene on chromosome 4, where theWheels (Whl) mutation has previously been mapped (Nolan et al.1995). The Whl-mutation is an ENU-induced mouse mutation that displays a complex phenotype, including hyperactive circling. The map location of the Eclb-gene on chromosome 4 (66 cM distal to the centromere) allowed us to rule out the possibility that the Eclb-gene is allelic to the Whl-mutation, which is localized within the 0–8 cM region distal to the centromere of chromosome 4 (Nolan et al.1995). Whirler (Lane 1963) andJerker (Eicher et al. 1977) are other candidate genes, localized in this region on chromosome 4. Both mouse mutants show circling behavior in addition to profound deafness. TheWhirler mutation is located 31.4 cM distal to the centromere (Lane 1963), and Jerker maps at 80.1 cM distal to the centromere (Eicher et al. 1977). Haplotypes of theEclb-candidate region must be constructed to indicate if one of these genes is located in the Eclb-candidate region.
In conclusion, the circling behavior in the Epistatic circlermouse is controlled by a necessary recessive Ecs-gene and a necessary Eclb-gene in combination with several facultative-modifying Ecl-genes. The unique inheritance pattern makes this model a valuable tool in the study of vestibular congenital malformation anomalies, because this trait in humans is in most cases a multifactorial condition as well, and unraveling the genetic basis will be much easier in an animal model. The four loci responsible for the circling phenotype in the Epistatic circler mouse are potential candidates for being involved in human genetic imbalance. Nevertheless, the actual role of these loci in vestibular dysfunction will remain obscure until further studies have been conducted and until the relation between the four genes is understood. This study illustrates the importance of animal models in the study of such complex diseases and at the same time provides evidence that identification of genetic determinants of multigenic traits is possible with modern molecular genetic tools.
METHODS
Mice
C57L/J and SWR/J mice were purchased from the Jackson Laboratory and IFFA CREDO (Charles River Company), respectively. Mouse care and experimental treatment were conducted in compliance with institutional and EU laws and policies.
Hearing Assessment
A handclap was used as a sound stimulus to detect mice with severe hearing impairment. A flick backward of the pinnae on hearing this sound was considered to be a positive Preyer reflex. We evaluated this reflex in all circling animals. Hearing in mice was also assessed by auditory brainstem response (ABR) measurements. Mice were anaesthetized with pentobarbital (60 mg/kg). Electrodes were placed subcutaneously near the ears. A Galileo Sirius system was used to deliver acoustic stimuli and measure the evoked brainstem response of anaesthetized mice. Mice were tested with broadband click stimuli at different intensities.
Isolation of DNA
A small piece of the tip of the tail of the mice was cut off using sharp scissors. The tail tip was incubated overnight at 37°C in a 655-μL lysis solution containing 10-mM Tris, 400-mM NaCl, 2-mM EDTA-di-Na, 1-μg/μL proteinase K, and 0.6% SDS. The solution was phenol/chloroform extracted and genomic DNA precipitated with ethanol. After redissolving the DNA, DNA concentrations were determined spectrophotometrically and samples were diluted to 100-ng/μL. For DNA samples used in the DNA-pooling experiment, a second spectrophotometric reading was taken to confirm the concentration.
Genetic Markers
For the genome scan, the Mouse MAPPAIRS Genome-Wide Screening Set was purchased from Research Genetics. This set contains 410 selected markers (developed by the Whitehead/MIT Center for Genome Research) with an average intermarker distance of 3.5 cM.
Before PCR, forward primers were radioactively labeled with [γ32P] ATP using T4 polynucleotide kinase. PCR reaction mixtures included 100-ng genomic DNA, 0.15-μM of each primer, 275-μM dNTPs, 0.35-U Taq polymerase, and 1.3-μL 10X PCR buffer containing 2-M Tris at pH 8, 3-M KCl, 100-mM MgCl2, 1% (v/v) Triton X-100, and 0.1% (w/v) gelatin. PCR conditions were as follows: initial denaturation of 3 min at 92°C was followed by 25 cycles of 15 sec at 92°C, 1 min at 55°C, and 1 min at 72°C. A final extension step consisted of 7 min at 72°C. The denaturated PCR products were run on a 6% polyacrylamide, 8-M urea-sequencing gel. Polymorphic bands were visualized by autoradiography. Extra markers for regions of interest were chosen from Mouse MAPPAIRS Genome-Wide Screening Set or from the Whitehead/MIT map (http://carbon.wi.mit.edu:8000/cgi-bin/mouse) if no suitable markers were available from the screening set. The extra markers wereD3Mit22, D3Mit142, D4Mit148,D4Mit251, D13Mit186, and D13Mit213. For markers that were not present in the Mouse MAPPAIRS Genome-Wide Screening Set, one of the primers was designed with an M13 sequence at the 5′end. A 5′-IRD (800 nm) M13 primer was included in the PCR reaction, thus labeling the PCR product. Gel electrophoresis and pattern visualization was performed using an LI-COR model 4200 DNA analyzer (NEN). For primers with an M13 sequence, different PCR conditions were used: PCR reaction mixtures included 100-ng genomic DNA, 0.05-μM of each primer, 170-μM dNTPs, 0.43-μM M13 primer, 0.35-U Taq polymerase, and 1-μl 10X PCR buffer. PCR conditions were as follows: initial denaturation of 3 min at 95°C was followed by 40 cycles of 1 min at 95°C, 1 min at 58°C, and 1 min at 72°C. A final extension step consisted of 20 min at 72°C.
Linkage Analysis
MLINK two-point linkage analysis using the LINKAGEsoftware package version 5.1 (Lathrop and Lalouel 1984) was performed between the Ecs-gene and the markers that suggested linkage, based on the results of the homozygosity mapping strategy. The frequency of the Ecs-gene was set at 0.0001. Equal recombination frequencies between males and females were assumed. LOD scores of 3.3 were considered as significant linkage (Lander and Kruglyak 1995). Because the LINKAGE software has limitations for the number of sibs in a pedigree, we were not able to calculate LOD score values for the genome scan for theEcl-genes. We therefore calculated χ-square values based on observed and expected values for the three genotypes (homozygous for the allele from C57L/J, heterozygous and homozygous for the allele from SWR/J). The expected values were based on the theoretically expected frequencies (1:2:1) for the three genotypes. P-values <10−4 were considered as significant for a genome-wide search (Lander and Kruglyak 1995).
Acknowledgments
We thank Peter Demant for helpful discussion regarding the statistical analysis and liability scale and Rudy D'Hooge for help with ABR measurements. This research was supported by the Life Sciences Foundation (NWO-ALW/SLW; project #805.33.313; A.M.V.A., CIDZ) and the Medical Research Division of The Dutch Organisation for Scientific Research (NWO-MW; CIDZ), and HFSP (CIDZ). K.C. holds a predoctoral position with the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT), and G.V.C. holds a research position with the FWO.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵5 Corresponding author.
-
E-MAIL gvcamp{at}uia.ua.ac.be; FAX 323 820 2566.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.218402.
-
Article published online before print in March 2002.
-
- Received October 11, 2001.
- Accepted January 29, 2002.
- Cold Spring Harbor Laboratory Press