The Human ATP-Binding Cassette (ABC) Transporter Superfamily
Abstract
The ATP-binding cassette (ABC) transporter superfamily contains membrane proteins that translocate a variety of substrates across extra- and intra-cellular membranes. Genetic variation in these genes is the cause of or contributor to a wide variety of human disorders with Mendelian and complex inheritance, including cystic fibrosis, neurological disease, retinal degeneration, cholesterol and bile transport defects, anemia, and drug response. Conservation of the ATP-binding domains of these genes has allowed the identification of new members of the superfamily based on nucleotide and protein sequence homology. Phylogenetic analysis is used to divide all 48 known ABC transporters into seven distinct subfamilies of proteins. For each gene, the precise map location on human chromosomes, expression data, and localization within the superfamily has been determined. These data allow predictions to be made as to potential functions or disease phenotypes associated with each protein. In this paper, we review the current state of knowledge on all human ABC genes in inherited disease and drug resistance. In addition, the availability of the completeDrosophila genome sequence allows the comparison of the known human ABC genes with those in the fly genome. The combined data enable an evolutionary analysis of the superfamily. Complete characterization of all ABC from the human genome and from model organisms will lead to important insights into the physiology and the molecular basis of many human disorders.
ABC Protein and Gene Organization
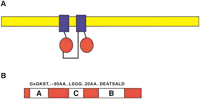
The ABC genes represent the largest family of transmembrane proteins. These proteins bind ATP and use the energy to drive the transport of various molecules across all cell membranes (Higgins 1992; Childs and Ling 1994; Dean and Allikmets 1995). Proteins are classified as ABC transporters based on the sequence and organization of their ATP-binding domains, also known as nucleotide-binding folds NBFs. The NBFs contain characteristic motifs, Walker A and B, separated by ∼90–120 amino acids, found in all ATP-binding proteins (Fig. 1). ABC genes also contain an additional element, the signature C motif, located just upstream of the Walker B site (Hyde et al. 1990). The functional protein typically contains two NBFs and two transmembrane (TM) domains (Fig. 1). The TM domains contain 6–11 membrane-spanning α-helices and provide the specificity for the substrate. The NBFs are located in the cytoplasm and transfer the energy to transport the substrate across the membrane. ABC pumps are mostly unidirectional. In bacteria, they are predominantly involved in the import of essential compounds that cannot be obtained by diffusion (e.g., sugars, vitamins, metal ions, etc.) into the cell. In eukaryotes, most ABC genes move compounds from the cytoplasm to the outside of the cell or into an extracellular compartment (endoplasmic reticulum, mitochondria, peroxisome). Most of the known functions of eukaryotic ABC transporters involve the shuttling of hydrophobic compounds either within the cell as part of a metabolic process or outside the cell for transport to other organs, or secretion from the body.
Diagram of a typical ABC transporter protein. (A) A diagram of the structure of a representative ABC protein is shown with a lipid bilayer in yellow, the transmembrane domains in blue, and the nucleotide binding fold in red. Although the most common arrangement is a full-transporter with motifs arranged N-TM-NBF-TM-NBF-C, as shown, NBF-TM-NBF-TM, TM-NBF, and NBF-TM arrangements are also found. (B) The NBF of an ABC gene contains the Walker A and B motifs found in all ATP-binding proteins. In addition, a signature or C motif is also present. The most common amino acids found in these motifs are shown above the diagram; subfamilies often contain characteristic residues in these and other regions.
The eukaryotic ABC genes are organized either as full transporters containing two TMs and two NBFs, or as half transporters (Hyde et al. 1990). The latter must form either homodimers or heterodimers to constitute a functional transporter. ABC genes are dispersed widely in eukaryotic genomes and are highly conserved between species, indicating that most of these genes have existed since the beginning of eukaryotic evolution. The genes can be divided into subfamilies based on similarity in gene structure (half vs. full transporters), order of the domains, and on sequence homology in the NBF and TM domains. There are seven mammalian ABC gene subfamilies, five of which are found in theSaccharomyces cerevisiae genome.
Overview of Human ABC Gene Subfamilies
A list of all known human ABC genes is displayed in Table1. This list includes an analysis of recently released genome sequences (Lander et al. 2001; Venter et al. 2001). Several sequences remain in the genome with homology to ABC genes, many of which represent pseudogenes. A comprehensive description of all ABC genes goes beyond the scope of this review. Therefore, only a concise summary of each subfamily is provided.
List of Human ABC Genes, Chromosomal Location, and Function
ABCA (ABC1)
This subfamily comprises 12 full transporters (Table 1) that are further divided into two subgroups based on phylogenetic analysis and intron structure (Broccardo et al. 1999). The first group includes seven genes dispersed on six different chromosomes (ABCA1, ABCA2, ABCA3, ABCA4, ABCA7, ABCA12, ABCA13), whereas the second group contains five genes (ABCA5, ABCA6, ABCA8, ABCA9, and ABCA10) arranged in a cluster on chromosome 17q24. The ABCA subfamily contains some of the largest ABC genes, several of which are >2100 amino acids. Two members of this subfamily, the ABCA1 and ABCA4 (ABCR) proteins, have been studied extensively. The ABCA1 protein is involved in disorders of cholesterol transport and high-density lipoproteins (HDL) biosynthesis (see below). The ABCA4 protein transports vitamin A derivatives in the outer segments of photoreceptor cells and therefore performs a crucial step in the visual cycle.
ABCB (MDR/TAP)
The ABCB subfamily is unique in that it contains both full transporters and half transporters. Four full transporters and seven half transporters are currently identified as members of this subfamily. ABCB1 (MDR/PGY1) is the first human ABC transporter cloned and characterized through its ability to confer a multidrug resistance phenotype to cancer cells. The functional sites of ABCB1 include the blood–brain barrier and the liver. The ABCB4 and ABCB11 proteins are both located in the liver and are involved in the secretion of bile acids. The ABCB2 and ABCB3 (TAP) genes are half transporters that form a heterodimer to transport peptides into the endoplasmic reticulum, which are presented as antigens by the Class I HLA molecules. The closest homolog of the TAPs, the ABCB9 half transporter, has been localized to lysosomes. The remaining four half-transporters, ABCB6, ABCB7, ABCB8 and ABCB10 localize to the mitochondria, where they function in iron metabolism and transport of Fe/S protein precursors.
ABCC (CFTR/MRP)
The ABCC subfamily contains 12 full transporters with a diverse functional spectrum that includes ion transport, cell surface receptor, and toxin secretion activities. The CFTR protein is a chloride ion channel that has a role in all exocrine secretions, and mutations in CFTR cause cystic fibrosis (Quinton 1999). ABCC8 and ABCC9 proteins bind sulfonylurea and regulate potassium channels involved in modulating insulin secretion. The rest of the subfamily is composed of nine MRP-related genes. Of these ABCC1, ABCC2, and ABCC3 transport drug conjugates to glutathione and other organic anions. The ABCC4, ABCC5, ABCC11 and ABCC12 proteins are smaller than the other MRP1-like genes and lack an animo-terminal domain (Borst et al. 2000) that is not essential for transport function (Bakos et al. 2000). The ABCC4 and ABCC5 proteins confer resistance to nucleosides including PMEA and purine analogs.
ABCD (ALD)
The ABCD subfamily contains four genes in the human genome and two each in the Drosophila and yeast genomes. The yeast PXA1 and PXA2 products dimerize to form a functional transporter involved in very long chain fatty acid oxidation in the peroxisome (Shani and Valle 1998). All of the genes encode half transporters that are located in the peroxisome, where they function as homo- and/or heterodimers in the regulation of very long chain fatty acid transport.
ABCE (OABP) and ABCF (GCN20)
The ABCE and ABCF subfamilies contain genes that have ATP-binding domains that are clearly derived from ABC transporters but have no TM domain and are not known to be involved in any membrane transport functions. The ABCE subfamily is comprised solely of the oligo-adenylate binding protein, a molecule that recognizes oligo-adenylate that is produced in response to infection by certain viruses. This gene is found in multicellular eukaryotes, but not in yeast, suggesting it is part of innate immunity. Each ABCF gene contains a pair of NBFs. The best-characterized member, the S. cerevisiae GCN20 gene mediates the activation of the eIF-2 α-kinase (Marton et al. 1997) and a human homolog, ABCF1, is associated with the ribosome and appears to have a similar role (Tyzack et al. 2000).
ABCG (White)
The human ABCG subfamily is comprised of six ‘reverse’ half transporters that have an NBF at the amino terminus and a TM domain at the carboxyl terminus. The most intensively studied ABCG gene is the white locus of Drosophila. The white protein, along with brown and scarlet, transport precursors of eye pigments (guanine and tryptophan) in the eye cells of the fly (Chen et al. 1996). The mammalian ABCG1 gene is involved in cholesterol transport regulation (Klucken et al. 2000). Other ABCG genes includeABCG2, a drug resistance gene; ABCG5 andABCG8, transporters of sterols in the intestine and liver;ABCG3, to date found exclusively in rodents; and theABCG4 gene that is expressed predominantly in the liver. The functions of the last two genes are unknown.
ABC Genes and Human Genetic Disease
Many ABC genes were originally discovered during the positional cloning of human genetic disease genes. To date, 14 ABC genes have been linked to disorders displaying Mendelian inheritance (Klein et al. 1999) (Table 2). As expected from the diverse functional roles of ABC genes, the genetic deficiencies that they cause also vary widely. Because ABC genes typically encode structural proteins, all of the disorders are recessive, and are attributable to a severe reduction or lack of function of the protein. Heterozygous variants in ABC gene mutations, however, are being implicated in the susceptibility to specific complex disorders.
Diseases and Phenotyes Caused by ABC Genes
Cystic Fibrosis and CFTR
Cystic fibrosis is the most common fatal childhood disease in Caucasian populations, reaching frequencies ranging from 1:900 to 1:2500. This corresponds to a carrier frequency of 1:15–1:25. The disease is much less common in African and Asian populations, where carrier frequencies of 1:100 to 1:200 have been estimated. The disease frequency correlates with the frequency of the major allele of the CF gene, a deletion of three base pairs (ΔF508). At least two other populations, however, have high frequency CF alleles. The W1282X allele is found on 51% of the alleles in the Ashkenazi Jewish population and the 1677delTA allele has been found at a high frequency in Georgians and is also present at elevated level in Turkish and Bulgarian populations. This has led several groups to hypothesize that these alleles arose through selection of an advantageous phenotype in the heterozygotes. It is through CFTR that some bacterial toxins such as cholera and Escherichia colicause increased fluid flow in the intestine and result in diarrhea. Therefore, several researchers have proposed that the CF mutations have been selected for in response to these diseases. This hypothesis is supported by studies showing that CF homozygotes fail to secrete chloride ions in response to a variety of stimulants, and a study in mice in which heterozygous null animals showed reduced intestinal fluid secretion in response to cholera toxin (Gabriel et al. 1993). CFTR is also the receptor for Salmonella typhimurium and implication in the innate immunity to Pseudomonas aeruginosa (Pier et al. 1998).
Patients with two severe CFTR alleles like ΔF508 typically display severe diseases with inadequate secretion of pancreatic enzymes leading to nutritional deficiencies, bacterial infections of the lung, and obstruction of the vas deferens, leading to male infertility. Patients with at least one partially functional allele display enough residual pancreatic function to avoid the major nutritional and intestinal deficiencies (Dean et al. 1990) and subjects with very mild alleles display only congenital absence of the vas deferens with none of the other symptoms of CF. Recently, heterozygotes of CF mutations have been found to have an increased frequency of pancreatitis (Cohn et al. 1998) and bronchiectasis (Pignatti et al. 1995). Therefore, there is a spectrum of severity in the phenotypes caused by this gene that is inversely related to the level of CFTR activity. Clearly, other modifying genes and the environment also affect disease severity, particularly the pulmonary phenotypes.
Adrenoleukodystrophy
Adrenoleukodystrophy (ALD) is an X-linked recessive disorder characterized by neurodegenerative phenotypes with onset typically in late childhood (Mosser et al. 1993). Adrenal deficiency commonly occurs and the presentation of ALD is highly variable. AMN, childhood ALD and adult onset forms are recognized, but there is no apparent correlation to ABCD1 alleles. ALD patients have an accumulation of unbranched, saturated fatty acids with a chain length of 24–30 carbons, in the cholesterol esters of the brain and in adrenal cortex. The ALD protein is located in the peroxisome, where it is believed to be involved in the transport of very long chain fatty acids.
Sulfonylurea Receptor
The ABCC8 gene is a high-affinity receptor for the drug sulfonylurea. Sulfonylureas are a class of drugs widely used to increase insulin secretion in patients with non-insulin-dependent diabetes. These drugs bind to the ABCC8 protein and inhibit an associated potassium channel. Familial persistent hyperinsulinemic hypoglycemia of infancy is an autosomal recessive disorder in which subjects display unregulated insulin secretion. The disease was mapped to 11p15-p14 by linkage analysis, and mutations in the ABCC8gene are found in PHHI families (Thomas et al. 1995). TheABCC8 gene has also been implicated in insulin response in Mexican-American subjects (Goksel et al. 1998) and in type II diabetes in French Canadians (Reis et al. 2000) but not in a Scandinavian cohort (Altshuler et al. 2000).
Bile Salt Transport Disorders
Several ABC transporters are specifically expressed in the liver, have a role in the secretion of components of the bile, and are responsible for several forms of progressive familial intrahepatic cholestasis (PFIC). PFICs are a heterogeneous group of autosomal recessive liver disorders, characterized by early onset of cholestasis that leads to liver cirrhosis and failure (Alonso et al. 1994). TheABCB4 (PGY3) gene transports phosphatidylcholine across the canalicular membrane of hepatocytes (van Helvoort et al. 1996). Mutations in this gene cause PFIC3 (Deleuze et al. 1996; de Vree et al. 1998) and are associated with intrahepatic cholestasis of pregnancy (Dixon et al. 2000). The rat Abcc2 gene was found to have a frame-shift mutation in the strain defective in canalicular multispecific organic anion transport, the TR− rat (Paulusma et al. 1996). The TR− rat is an animal model of Dubin-Johnson syndrome and mutations in ABCC2 have been identified in Dubin-Johnson syndrome patients (Wada et al. 1998). The ABCC2 protein is expressed on the canalicular side of the hepatocyte and mediates organic anion transport. The ABCB11 gene was originally identified based on homology to ABCB1 (Childs et al. 1995).ABCB11 is highly expressed on the liver canalicular membrane and has been shown to be the major bile salt export pump. Mutations inABCB11 are found in patients with PFIC2 (Strautnieks et al. 1998).
Retinal Degeneration and ABCA4
The ABCA4 gene is expressed exclusively in photoreceptors where it transports retinol (vitamin A) derivatives from the photoreceptor outer segment disks into the cytoplasm (Allikmets et al. 1997). The chromophore of a visual pigment rhodopsin, retinal, or conjugates with phospholipids are the likely substrates for ABCA4, as they stimulate the ATP hydrolysis of the protein (Sun et al. 1999). Mice lacking Abca4 show increased all-trans-retinaldehyde (all-trans-RAL) following light exposure, elevated phosphatidylethanolamine (PE) in outer segments, accumulation of the protonated Schiff base complex of all-trans-RAL and PE (N-retinylidene-PE), and striking deposition of a major lipofuscin fluorophore (A2-E) in retinal pigment epithelium (RPE) (Weng et al. 1999). These data suggest that ABCR is an outwardly directed flippase for N-retinylidene-PE.
Mutations in the ABCA4 gene have been associated with multiple eye disorders (Allikmets 2000). A complete loss of ABCA4 function leads to retinitis pigmentosa, whereas patients with at least one missense allele have Startgardt disease (STGD). STGD is characterized by juvenile to early adult onset macular dystrophy with loss of central vision. ABCA4 mutation carriers are also increased in frequency in age-related macular degeneration (AMD) patients. AMD patients display a variety of phenotypic features, including the loss of central vision after the age of 60. The causes of this complex trait are poorly understood, but a combination of genetic and environmental factors have a role. The abnormal accumulation of retinoids, caused by ABCA4 deficiency has been postulated to be one mechanism by which this process could be initiated. Defects in ABCA4 lead to an accumulation of retinal derivatives in the retinal pigment epithelium behind the retina.
Mitochondrial Iron Homeostasis
Several half transporters of the MDR/TAP subfamily have been localized to the inner membrane of the mitochondria. The yeast ortholog of ABCB7, Atm1, has been implicated in mitochondrial iron homeostasis, as a transporter in the biogenesis of cytosolic Fe/S proteins (Kispal et al. 1997). Two distinct missense mutations in ABCB7 are associated with the X-linked sideroblastic anemia and ataxia (XLSA/A) phenotype (Allikmets et al. 1999). Three more half transporters from this subfamily, ABCB6, ABCB8 and ABCB10 have also been localized to mitochondria (Table 1).
Sterol Transport Deficiencies
Tangier disease is characterized by deficient efflux of lipids from peripheral cells, such as macrophages, and a very low level of HDL. The disease is caused by alterations in the ABCA1 gene, implicating this protein in the pathway of removal of cholesterol and phospholipids onto HDL (Young and Fielding 1999). Patients with hypolipidemia have also been described that are heterozygous forABCA1 mutations, suggesting that ABCA1 variations may have a role in regulating the level of HDLs in the blood (Marcil et al. 1999).
Subsequently, the sterol-dependent regulation of ABCA1expression was shown (Langmann et al. 1999). Current models for ABCA1 function place it at the plasma membrane where it mediates the transfer of phospholipid and cholesterol onto lipid-poor apolipoproteins to form nascent HDL particles. The ABCA1-mediated efflux of cholesterol is regulated by nuclear hormone receptors, such as oxysterol receptors (LXRs) and the bile acid receptor (FXR), as heterodimers with retinoid X receptors (RXRs) (Repa et al. 2000).
Recently, two half-transporter genes, ABCG5 and ABCG8were characterized (Berge et al. 2000; Lee et al. 2001), located head-to-head on the human chromosome 2p15-p16, and regulated by the same promoter. These genes are both mutated in families with sitosterolemia, a disorder characterized by defective transport of plant and fish sterols and cholesterol. Most likely, the two half-transporters form a functional heterodimer. The ABCG1gene is also regulated by cholesterol (Klucken et al. 2000) andABCG4 is highly expressed in the liver, suggesting that these two genes may also be involved in cholesterol transport (Table 1).
Multidrug Resistance
Cells exposed to toxic compounds can develop resistance by a number of mechanisms including decreased uptake, increased detoxification, alteration of target proteins, or increased excretion. Several of these pathways can lead to multidrug resistance (MDR), in which the cell is resistant to several drugs in addition to the initial compound. This is a particular limitation to cancer chemotherapy and the MDR cell often displays other properties, such as genome instability and loss of checkpoint control, which complicate further therapy. ABC genes have an important role in MDR and at least six genes are associated with drug transport (Table 3).
ABC Transporters Involved in Drug Resistance
ABCB1
The best characterized ABC drug pump is the ABCB1 gene, formerly known as MDR1 or PGY1. ABCB1 was the first human ABC transporter cloned and characterized through its ability to confer a multidrug resistance phenotype to cancer cells that had developed resistance to chemotherapy drugs (Juliano and Ling 1976). ABCB1 has been demonstrated to be a promiscuous transporter of hydrophobic substrates, hydrophobic drugs including drugs including colchicine, VP16, adriamycin and vinblastine as well as lipids, steroids, xenobiotics, and peptides (for review, see Ambudkar 1998). The gene is thought to have an important role in removing toxic metabolites from cells, but is also expressed in cells at the blood–brain barrier and presumably has a role in transporting compounds into the brain that cannot be delivered by diffusion. ABCB1 also affects the pharmacology of the drugs that are substrates and a common polymorphism in the gene affects digoxin uptake (Hoffmeyer et al. 2000).
ABCC1
The ABCC1 gene was identified in the small-cell lung carcinoma cell line NCI-H69, a multidrug resistant cell that did not overexpress ABCB1 (Cole et al. 1992). The ABCC1 pump confers resistance to doxorubicin, daunorubicin, vincristine, colchicines and several other compounds, a very similar profile to that of ABCB1. Unlike ABCB1, however, ABCC1 transports drugs that are conjugated to glutathione by the glutathione reductase pathway (Borst et al. 2000). ABCC1 can also transport leukotrienes, such as leukotriene C4 (LTC4). LTC4 is an important signaling molecule for the migration of dendritic cells. Migration of dendritic cells from the epidermis to lymphatic vessels is defective in Abcc1 −/− mice (Robbiani et al. 2000).
ABCG2
Analysis of cell lines resistant to mitoxantrone that do not overexpress ABCB1 or ABCC1, led several laboratories to identify theABCG2 (ABCP, MXR1, BCRP) gene as a drug transporter (Allikmets et al. 1998; Doyle et al. 1998; Miyake et al. 1999). ABCG2 confers resistance to anthracycline anticancer drugs and is amplified or involved in chromosomal translocations in cell lines selected with topotecan, mitoxantrone, or doxorubicin treatment. It is suspected that ABCG2 functions as a homodimer because tranfection of the gene into cells confers resistance to chemotherapeutic drugs. ABCG2 can also transport several dyes such as rhodamine and Hoechst 33462 and the gene is highly expressed in a subpopulation of hematopoetic stem cells (side population) that stain poorly for these dyes. The normal function of the gene in these cells, however, is unknown. ABCG2 is highly expressed in the trophoblast cells of the placenta. This suggests that the pump is responsible either for transporting compounds into the fetal blood supply, or removing toxic metabolites. The gene is also expressed in the intestine and ABCG2 inhibitors that could be useful in making substrates orally available.
Phylogenetic Analysis of Human ABC Genes
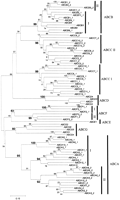
The identification of the nearly complete set of human ABC genes allows a comprehensive phylogenetic analysis of the superfamily. To understand the organization of these genes within the subfamilies described previously, an alignment of the ATP-binding domains was generated and used for phylogenetic analysis. Figure2 displays a neighbor-joining tree resulting from this analysis. The proposed nomenclature of ABC transporters is in excellent agreement with the phylogenetic trees obtained. In particular, all major ABC transporter families are represented in the human tree by stable clusters with high bootstrap values.
Phylogenetic tree of the human ABC genes. ATP-binding domain proteins were identified using the model ABC_tran (accession PF00005) of the pfam database (Bateman et al. 1999). The HMMSEARCH program from the HMMER package (Eddy 1998) and a set of custom-made service scripts were used to extract ATP-binding domains from all protein sequences of interest. Note that some proteins analyzed contain two ATP-binding domains (denoted on the figures as I and II), whereas others contained only one ATP-binding domain. Alignments were generated with the hidden Markov model (Eddy 1995) based on theHMMALIGN program using the ABC_tran model. The resulting multiple alignment was analyzed with NJBOOT (N. Takezaki, pers. comm.) implementing the neighbor-joining tree-making algorithm (Saitou and Nei 1987) and the number at the branch of the nodes represents the value from 100 replications. The distance measure between sequences used for tree making was the Poisson correction for multiple hits (Zuckerkandl and Pauling 1965). To verify the position of the previously unknown subgroup of Drosophila genes (CG6162, CG6162, and CG11147), they were aligned with a representative of each of the human subfamilies. Because some of the human proteins had two ATP-binding domains, the set contained three Drosophila and 12 human sequences. The JTT (Jones et al. 1992) model as defined in the MOLPHY package with the ‘star decomposition’ option was employed. The tentative best tree (the total number of possible trees for 15 sequences is too large for exhaustive search through all these trees) was then used for local maximum likelihood search through the surrounding tree topologies.
This analysis provides compelling evidence for frequent domain duplication of ATP-binding domains in ABC transporters. Virtually invariably, both ATP-binding domains within a gene are more closely related to each other than to ATP-binding domains from ABC transporter genes of other subfamilies. This could be explained by a concerted evolution of domains within the same gene, but this seems unlikely because the two domains within each gene are substantially diverged. Therefore, it appears that duplication of ATP-binding domains within major ABC families was attributable to several independent duplication events rather than a single ancestral duplication.
Drosophila ABC genes
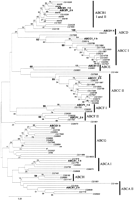
To begin to understand the organization and evolution of theDrosophila ABC genes, the Celera (Myers et al. 2000) and FlyBase databases were searched for sequences by a combination ofBLAST searches and analysis of the annotation already present in the databases. Initial subfamily classifications were assigned based on homology and BLAST scores, and the location of each gene was recorded (Table4). In total, 56 genes were identified and there is at least one representative of each of the known mammalian subfamilies (Table 5). To confirm the subfamily groupings, the ATP-binding domain amino acid sequences were used to perform phylogenetic analyses. A representative tree is shown in Figure 3. As expected, genes from the same subfamily cluster together and confirm the initial assignments made by inspection.
Drosophila ABC Genes
ABC Gene Subfamilies in Characterized Eukaryotes
Phylogenetic tree of the Drosophila ABC genes. Analysis (see Fig. 2) was performed with all extracted Drosophila sequences and a representative of each human subfamily.
As in the human and yeast genomes, the Drosophila ABC genes are largely dispersed in the genome. There are four clusters of two genes and one cluster of four genes (Table 4). One of these clusters (on chromosome 2L, band 37B9) is composed of an ABCB and an ABCC gene indicating that this is a chance grouping of genes. The remaining clusters are composed of genes from the same subfamily and arranged in a head-to-tail fashion consistent with gene duplication. Because the clusters are themselves dispersed and involve different subfamilies, they presumably represent independent gene duplication events.
The best-studied Drosophila ABC genes are the eye pigment precursor transporters white, scarlet, and brown (w,st, and br, respectively). These genes are part of the ABCG subfamily and have a unique NBF–TM organization. Surprisingly, there are 15 ABCG genes in the fly genome, making this the most abundant ABC subfamily. This is in sharp contrast to the only five and six known ABCG genes in the human and mouse genomes, respectively. The Drosophila ABCG genes are highly dispersed in the genome with only two pairs of linked genes. In addition, they are quite divergent phylogenetically, suggesting that there were many independent and ancient gene duplication events.
Several Drosophila ABCB genes, Mdr49, Mdr50, and Mdr65, have also been well characterized. A fourth member of this group, CG10226, was identified that is clustered withMdr65 (Table 4). These genes are closely related to the human and mouse P-glycoproteins (ABCB1, ABCB4) and disruption of Mdr49 results in sensitivity to colchicines (Wu et al. 1991).
To search for potential gene functions, each region containing aDrosophila ABC gene was searched for phenotypes that have been not assigned to a gene (Table 4). The most promising connection is the identification of several eye phenotypes (vin, rose, and cln) in the region of the CG7346 gene. BecauseCG7346 is part of the ABCG family and is therefore related tow, st, and br, it is tempting to speculate that mutations in CG7346 cause one or more of these phenotypes. Because ABC genes perform very diverse functions and are associated with varied phenotypes it is hard to gather much additional insight from this analysis.
Three genes, CG9990, CG6162, and CG11147, were identified that do not fit into any of the known subfamilies and, in fact, are most closely related to ABC genes from bacteria. There are no close homologs to these genes in any other eukaryotic genome including worm and plants. These genes are within large contiguous sequences and have introns, therefore they do not represent contamination from bacterial sequences. This group forms a distinct cluster on the Drosophila tree. To elucidate the position of this group with respect to human ABC transporter families, we produced two datasets with nine and 15 sequences, respectively, containing ATP-binding domains from the three Drosophila genes and from one representative from each of the human families. In the smaller subset, only one ATP-binding domain from each human gene was used. These two datasets were subjected to maximum likelihood analysis, with a heuristic algorithm for the larger dataset and with a rigorous exhaustive search for the smaller datasets. The result of the analysis clearly indicates that the new subfamily of ABC transporter inDrosophila is significantly different from all known families of ABC transporters (data not shown) and might have a yet unidentified functional role. We propose designating this new group of genes as subfamily H. DNA sequence searches showed that although significant homology is present between these three genes in both the NBF and TM regions, there is no homology in the TM regions with any other eukaryotic or prokaryotic ABC proteins. The closest related NBF sequences are all bacterial, such as the E. coli YHIH gene, a ribosomal ATPase. This gene shows 36% amino acid identity in the NBF domain.
The ATP-binding domains in a generic ABC transporter appear to be following a birth-and-death process that has been described before for other multigene families. In a typical birth-and-death process (Ota and Nei 1994; Nei et al. 1997), repeated genetic segments (in this case domains) experience nearly random fluctuation in their number. That is, genomic deletions decrease segment number, while genome duplications increase it. The phylogenetic trees that we present here for human andDrosophila give clear evidence of the birth-and-death evolution of ATP-binding domains in ABC transporters, and similar analyses of the yeast genes support this. In S. cerevisiae, there are several G subfamily ABC genes (e.g., YOR011w and SNQ2) that have two NBF and TM domains (Michaelis and Berkower 1995; Decottignies and Goffeau 1997). In contrast, all of the Drosophila and human ABCG family genes are half transporters. Therefore, it appears that the second ATP-binding domain in animal ABCG genes was lost in an ancestral lineage preceding the animal radiation but not the animal-fungi split. Similarly, ABCE genes in humans are one-domain genes, whereas both Drosophila and yeast have two-domain homologs (RLI1 in yeast and CG5651 in Drosophila). Therefore, the loss of the second domain appears to be a relatively recent event in human ABCE genes, probably in the vertebrate lineage.
Perspectives
One of the most fascinating findings from the analysis of the ABC genes from the human, worm, and fly genomes is the remarkable similarity in the number of ABC genes. Although additional sequencing and annotation will undoubtedly change the exact number somewhat, it is clear that humans do not have substantially more ABC genes than much simpler eukaryotes. This suggests that there is a core of essential ABC genes that are required for all multicellular eukaryotes. These higher eukaryotes do have about twice the number of ABC genes as does S. cerevisiae, suggesting that the evolution of multicellularity was accompanied by the expansion in the number of ABC genes. It is clear, however, that certain species have expanded some subfamilies more than others. As mentioned above, flies have considerably more ABCG genes than humans (15 vs. five), whereas there are at least 12 ABCA genes in the human and only 10 in Drosophila. Despite the remarkable similarity in gene number, most Drosophila ABC genes do not have a clear ortholog in the human genome. The likely exceptions are the ABCD, ABCE, and ABCF subfamilies that contain nearly identical gene numbers in the two species. The only ABC genes in the human and yeast genomes that are documented to have similar function are the ABCD (ALD-like) genes that are involved in the transport of very long chain fatty acids, and the ATM1/ABCB7 gene that is present in the mitochondria and has a role in iron metabolism.
A major limitation to the understanding of ABC genes is the difficulty in obtaining crystal structures. The three-dimentional structure of two bacterial NBF proteins has been obtained and has greatly improved our understanding of the organization and function of this portion of the protein (Hung et al. 1998). It is clear that conformational changes in the NBF influence substrate binding and transport, and to date, this can only be addressed by laborious mutagenesis and biochemical experiments (Hafkemeyer et al. 1998).
Ideally, the worm and fly ABC genes could be used to elucidate the function of their human counterparts. To date, however, we know considerably more about human ABC gene function that we do about either the Drosophila or Caenorhabditis elegans ABC genes. In fact, the only Drosophila genes initially identified based on their function are the eye pigment genes. Similarly, only one ABC gene in C. elegans (ced-7) was identified based on function (involvement in cell death; Wu and Horvitz 1998). It is not obvious why more ABC genes are not associated with visible or detectable phenotypes in Drosophila or C. elegans as many of the human ABC genes affect morphology, behavior, lifespan, and fertility and these are traits that have been observed and selected for repeatedly in these organisms. These model organisms, however, do provide an important resource for the future systematic study of ABC genes. Analysis of ABC gene expression combined with gene disruptions should yield important clues to gene function. In addition, the capability of designing suppressor screens may allow the identification of pathways that involve ABC genes.
Acknowledgments
We thank Kirby Smith for helpful comments on the manuscript and apologize to all whose primary papers could not be cited because of lack of space.
Footnotes
-
↵6 Corresponding author.
-
E-MAIL dean{at}ncifcrf.gov; FAX (301) 846-1909.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.184901.
- Cold Spring Harbor Laboratory Press