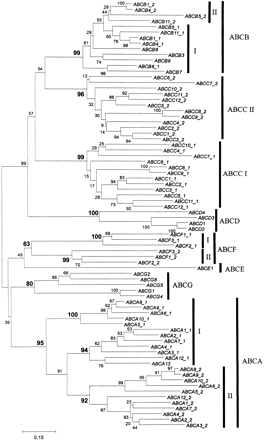
Phylogenetic tree of the human ABC genes. ATP-binding domain proteins were identified using the model ABC_tran (accession PF00005) of the pfam database (Bateman et al. 1999). The HMMSEARCH program from the HMMER package (Eddy 1998) and a set of custom-made service scripts were used to extract ATP-binding domains from all protein sequences of interest. Note that some proteins analyzed contain two ATP-binding domains (denoted on the figures as I and II), whereas others contained only one ATP-binding domain. Alignments were generated with the hidden Markov model (Eddy 1995) based on theHMMALIGN program using the ABC_tran model. The resulting multiple alignment was analyzed with NJBOOT (N. Takezaki, pers. comm.) implementing the neighbor-joining tree-making algorithm (Saitou and Nei 1987) and the number at the branch of the nodes represents the value from 100 replications. The distance measure between sequences used for tree making was the Poisson correction for multiple hits (Zuckerkandl and Pauling 1965). To verify the position of the previously unknown subgroup of Drosophila genes (CG6162, CG6162, and CG11147), they were aligned with a representative of each of the human subfamilies. Because some of the human proteins had two ATP-binding domains, the set contained three Drosophila and 12 human sequences. The JTT (Jones et al. 1992) model as defined in the MOLPHY package with the ‘star decomposition’ option was employed. The tentative best tree (the total number of possible trees for 15 sequences is too large for exhaustive search through all these trees) was then used for local maximum likelihood search through the surrounding tree topologies.











