Neuropeptides and Neuropeptide Receptors in the Drosophila melanogaster Genome
Abstract
Recent genetic analyses in worms, flies, and mammals illustrate the importance of bioactive peptides in controlling numerous complex behaviors, such as feeding and circadian locomotion. To pursue a comprehensive genetic analysis of bioactive peptide signaling, we have scanned the recently completed Drosophila genome sequence for G protein-coupled receptors sensitive to bioactive peptides (peptide GPCRs). Here we describe 44 genes that represent the vast majority, and perhaps all, of the peptide GPCRs encoded in the fly genome. We also scanned for genes encoding potential ligands and describe 22 bioactive peptide precursors. At least 32 Drosophila peptide receptors appear to have evolved from common ancestors of 15 monophyletic vertebrate GPCR subgroups (e.g., the ancestral gastrin/cholecystokinin receptor). Six pairs of receptors are paralogs, representing recent gene duplications. Together, these findings shed light on the evolutionary history of peptide GPCRs, and they provide a template for physiological and genetic analyses of peptide signaling in Drosophila.
The recent publication of entire genomes for the worm, the fly, and human species has initiated the era of functional genomic analysis. The experiences to date have indicated that such analysis involves multiple stages, in which improvements are recorded as the databases are completed and analytic programs become more precise (Reese et al. 2000), and as more comparative information is made available (Sonnhammer et al. 1997). G protein-coupled receptors (GPCRs) provide sensitivity to a variety of environmental, developmental, and physiological signals. They display a uniform topology with seven transmembrane (TM) domains and represent one of the largest recognizable groups of proteins (Bockaert and Pin 1999). Here we have organized all genomic sequences that encodeDrosophila GPCRs to identify and classify those devoted to peptide hormone and neuropeptide ligands (peptide GPCRs).
Given the availability of the human and mouse genomic sequences, what can we gain by a thorough analysis of Drosophila peptide GPCRs? We propose two reasons to motivate such efforts. First, our understanding of GPCR signaling mechanisms appears incomplete. Recent advances have indicated means by which GPCR signaling potential may be increased, including receptor oligomerization and association with a variety of accessory proteins (Bockaert and Pin 1999), and receptor translocation to the nucleus (Chen et al. 2000). There is great need, therefore, to address new hypotheses of GPCR signaling mechanisms in vivo. For this purpose, it will be very helpful to use the powerful tools for genetic analysis that are afforded by model organisms such asDrosophila. The second reason we favor the pursuit ofDrosophila GPCRs invokes the success of genetic analysis in another model genetic system, Caenorhabditis elegans. In the past few years, rapid progress has been made in the analysis of insulin signaling in the worm; C. elegans insulin regulates metabolism, development, and longevity by mechanisms that are similar to the endocrine regulation of metabolism and fertility by mammalian insulin (Kimura et al. 1997; Tissenbaum and Ruvkun 1998). In addition, the genetic analysis has extended our understanding of insulin signaling by revealing novel molecular features that may be variant in diabetic pedigrees (Ogg et al. 1997; Ogg and Ruvkun 1998). Although insulin binds to a different class of receptors, it is likely that the same rapid development of new information will accompany the genetic analysis of peptide GPCRs in Drosophila.
In a recent review, Brody and Cravchik (2000) began the process of categorizing Drosophila GPCRs by describing ∼100 genes, including 21 receptors for classical neurotransmitters and neuromodulators (biogenic amines, related compounds, and purines) and 26–30 peptide receptor genes. We have extended that analysis by re-searching the original genomic sequences for peptide receptors (we found one additional GPCR) and by improving the annotations of 20 previously-predicted genes. We classified receptor genes according to phylogenetic trees constructed with the aid of the Pfam 7 TM databases (Bateman et al. 2000). In addition, we refined the DrosophilaGPCR classifications by incorporating information we deduced by examining gene organizations. Through this analysis, we expanded the set of known and candidate peptide GPCRs from ∼30 to ∼45. Finally, to gain a sense of the potential peptide ligands, we assembled a list of 22 Drosophila genes known or predicted to encode bioactive peptides that may activate these receptors. Together, these results shed light on the evolutionary history of neuropeptide signaling. They also are intended to aid in future efforts to analyze peptide receptor function in development, physiology, and behavior by using the power of Drosophila genetics.
RESULTS AND DISCUSSION
We searched the Drosophila melanogaster genome sequence with the goal of identifying all peptide GPCRs. Initially, we scanned the gene annotations developed jointly by the BerkeleyDrosophila Genome Project (BDGP) and Celera Genomics for all putative GPCRs (Adams et al. 2000; Brody and Cravchik 2000). Based onBLASTP scores obtained with each sequence, we excluded entries that are likely to represent nonpeptide GPCRs (neurexins, HE6- and methuselah-related proteins, rhodopsins, developmental genes, taste and odorant receptors, and receptors for biogenic amines and other small neurotransmitters). The remaining set of 44 known or putative peptide receptors and unclassified GPCRs was retained for further analysis (Table 1).
Cloned and Candidate Drosophila Neuropeptide Receptors
To gauge the completeness of this set, we scanned theDrosophila genome for additional, nonannotated peptide GPCRs in three ways. First, we scanned a set of annotations of GPCR genes obtained through a GENSCAN search of the entireDrosophila genome (K. Scott and L. Vosshall, pers. comm.). This list contained one receptor sequence, BG:BACR48G21.1(BACR48G21.1), which had not been annotated previously. Second, we performed BLASTP searches using a “GPCR query set,” which included the previously cloned or annotated peptide, amine, and related/unclassified Drosophila GPCRs as well as ∼200 sequences representing a diverse set of Family A GPCRs (unless indicated, we used the nomenclature for GPCR Families and Groups given by Kolakowski [1994] [see http://www.gcrdb.uthscsa.edu/]). All of the putative peptide receptors listed in Table 1 (including BACR48G21.1) were detected with several queries (for the vast majority, more than 30 times). However, this analysis did not reveal any additional candidate peptide GPCRs. Finally, we used the GPCR query set to perform TBLASTN searches of the Celera/BDGP whole genome shotgun sequence. As with the BLASTP survey, theTBLASTN search yielded scaffold sequences corresponding to all of the GPCRs on our list, but no additional candidate peptide receptors. The whole genome shotgun sequence currently represents ∼98% of the Drosophila eukaryotic genome (Adams et al. 2000). Therefore, we conclude that the set of 45 cloned and candidate peptide GPCRs is essentially complete.
We focused on sequences encoding the seven TM domains. More than 50% of the BDGP/Celera annotations for GPCRs in this set (23 out of 43) were missing sequences representing one or more TM domains or, in the case of two receptors (CG4187 and CG5042), an N-terminal domain containing conserved, leucine-rich repeats (Table 1). In three cases, the correct gene sequences were published previously (Li et al. 1991;Ashburner et al. 1999; Birgül et al. 1999). We revised the remaining 20 incorrect or incomplete annotations using software-based gene prediction methods and manual inspection. For six GPCRs, there were two to three neighboring annotations that contained nonoverlapping GPCR sequence motifs. In each of these cases, we did not detect open reading frames encoding conserved TM domains in the intervening genomic sequence. Therefore, we merged these sequences to generate single, revised annotations. Additionally, for CG5911, we detected two adjacent sets of exons encoding alternative versions of TM4–7, including a conserved splice acceptor site in TM4. Thus, CG5911 appears to encode two distinct receptor isoforms (50% identical in TM5–7) through alternative splicing. Although final confirmation of these predictions will require direct sequencing of cDNAs, we conclude that our revised annotations are of sufficiently high quality to perform phylogenetic analysis, based on the presence of well conserved motifs (Baldwin et al. 1997; Tams et al. 1998) throughout the TM domains of each of these receptors.
After assembling the list of 45 cloned and candidate peptide GPCRs, we classified these proteins based on BLASTP scores, on the locations of these receptors on representative phylogenetic trees for each GPCR Family (A and B), and on the degree to which these locations were supported by bootstrap analysis. We examined the genomic location of each peptide GPCR as well as all biogenic amine and small transmitter GPCRs to identify linked (possibly paralogous) genes. To detect conserved gene organizations, we noted the intron locations and phasing for each cloned and candidate peptide GPCR within the TM regions, as well as for a few related vertebrate GPCRs. The intron analysis of the vertebrate GPCRs was not comprehensive, in part because >93% of vertebrate GPCR genes lack introns within the coding sequence (Gentles and Karlin 1999). Finally, based on the results of the above tests, we were able to place most of the Family A receptors in one of seven alignments, each of which contained one or more related receptor subgroups.
We found strong evidence supporting the classification of 32 receptors as peptide GPCRs (Table 2). In addition, there were two receptors that are clear orthologs of the orphan receptor, LGR7, a member of a receptor clade containing several peptide GPCRs. For seven additional receptors, we found weaker evidence to indicate that they are peptide GPCRs. Finally, we regard four of the receptors as unclassifiable. Interestingly, we found at least six pairs of paralogs (variants generated by gene duplications or other processes), most of which appear to be related to common ancestors of vertebrate GPCR subgroups (rather than derived independently from vertebrate paralogs; e.g., Fig. 1A,D [see below]). Based on the presence of ESTs and/or cDNAs (Table 1), at least 25% of the genes in Table 1 are expressed. Pseudogenes are rare in Drosophila(Petrov and Hartl 2000), and based on strong sequence conservation in the TM domains, most of the remaining genes are likely to be expressed as well. Thus, we conclude that there are 39–41 peptide GPCRs in Drosophila, with an additional four GPCRs that may later be included in this category. The following sections describe this analysis in detail and provide a listing of potential cognate ligands.
Classification of Cloned and Candidate DrosophilaNeuropeptide Receptors by BLAST and Phylogenetic Analysis
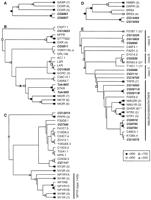
Neighbor-joining phylogenetic trees for the Family A, Group III-B receptors. For Family and Group classifications, see Kolakowski (1994) (see http://www.gcrdb.uthscsa.edu/). (A) Rooted tree for the gastrin/cholecystokinin (CCK) receptors. (B) Unrooted tree for the neurokinin receptors (NKRs) and related GPCRs. The midpoint of the tree is indicated with an “X.” (C)Unrooted tree for the neuropeptide Y (NPY) receptors (NPYRs) and prolactin releasing receptors (PRPRs). (D) Rooted tree for the bombesin/gastrin releasing peptide receptors. (E) Unrooted tree for the neuromedin U receptors (NMURs), growth hormone secretagogue receptors (GHSRs), neurotensin receptors (NTRs), thyrotropin releasing hormone receptors (TRFRs), and a large family of related orphan receptors from Caenorhabditis elegans. The C. elegans orphan receptors included here belong to one of three clades (classes A–C). (*) Omitted receptors are additional C. elegansorphan GPCRs; (**) omitted receptors are additional GHSRs and closely related orphan GPCRs. In A and D, a monophyletic set of 26 biogenic amine receptors (not shown) was used as the outgroup to determine the root of the trees (see Methods). In C andE, the location of the tree midpoint was ambiguous and is therefore not indicated. Portions of the trees representing groups of closely related receptors were omitted (the number of related receptors on each branch is indicated in parentheses). Drosophila GPCRs are listed in bold and italics. (BRS3) Bombesin receptor subtype 3; (BRS4) bombesin receptor subtype 4; (CCKR) CCK receptor type A; (CCKR XL) Xenopus laevis CCKR; (GASR) gastrin/CCK receptor type B; (GCRC) glucocorticoid-induced receptor; (GRL106) Lymnaea stagnalis cardioexcitatory receptor; (GRPR) gastrin releasing peptide (GRP) receptor; (LKR) Boophilus microplus (tick) leucokinin-like peptide receptor; (LSR) L. stagnalislymnokinin receptor; (NFFR) neuropeptide FF/neuropeptide AF receptor; (NK1R–NK3R) NKR types 1–3; (NMU1R and NMU2R) neuromedin U receptor types 1 and 2; (NPR-1) product of the C. elegans npr-1 gene; (NPYRYA–NPYRYC) orphan zebrafish NPYRs; (NPYRB) Gadus morhua(Atlantic cod) NPYR; (NTR1 and NTR2) neurotensin receptor types 1 and 2; (NY1R–NY6R) NPY receptor types 1–6; (OT7T022) putative mammalian RFRP receptor; (OXR) orexin/hypocretin receptor; (STKR) Stomoxys calcitrans (stable fly) tachykinin receptor. The remaining non-Drosophila sequences are orphan GPCRs from C. elegans. Symbols denote bootstrap support, out of 1000 replicates, that was >500: (filled circles) >990; (open circles) >900; (open squares) >700; (open triangles) >500.
Overview of the Drosophila Peptide GPCRs
Together, the set of known and candidate Drosophila peptide GPCRs contains representatives of at least 15 monophyletic vertebrate GPCR subgroups. Family A/Group III-B contains the largest number ofDrosophila peptide GPCRs (at least 19; Table 2). These include 17 Drosophila representatives of six vertebrate GPCR subgroups: the gastrin/cholecystokinin, neurokinin, neuropeptide FF and hypocretin/orexin, neuropeptide Y, bombesin/gastrin releasing peptide, and neurotensin receptors (and the neurotensin-related receptors for neuromedin U, growth hormone secretagogue, and thyrotropin releasing hormone). Family A/Group V also contained a large number ofDrosophila peptide GPCRs (11; Table 2), representing seven vertebrate GPCR subgroups: the galanin, somatostatin/opioid, gonadotropin releasing hormone, oxytocin/vasopressin, and glycoprotein hormone receptors, as well as two subgroups represented by vertebrate orphan receptors (LGR4–6 and LGR7). Finally, there were fiveDrosophila peptide GPCRs that belong to Family B. Four of these receptors belong to one of two vertebrate GPCR subgroups: the calcitonin and corticotropin releasing factor receptors. Thus, a large majority of the Drosophila and vertebrate neuropeptide signaling pathways appear to share common evolutionary origins. It remains to be seen whether the functions of these signals have been similarly conserved.
Family A/Group III-B: Gastrin/Cholecystokinin Receptors
Cholecystokinin (CCK) and gastrin are related neuroendocrine peptides that act through two closely related families of receptors (type A, CCKR, and type B, GASR). These receptors likely evolved from a common ancestor (Johnsen 1998). Two Drosophila GPCRs (CG6857, CG6881) displayed strong evidence of evolutionary kinship with this receptor subgroup (Table 2). On the subgroup-specific tree (Fig.1A), CG6857 and CG6881 (as well asXenopus laevis CCKR) were located on the base of the tree, before the branches leading to the CCKR and GASR receptors. Therefore, it appears that the fly receptors diverged from a common ancestor of the CCKR and GASR lineages. Consistent with this interpretation,CG6857 and CG6881 are closely linked genes (∼30 kb apart), and they each display the strongest sequence similarity with each other (by BLASTP and on the phylogenetic trees; Table2, Fig. 1A). Therefore, they likely arose through a gene duplication event rather than independently from the CCKR and/or GASR lineages.CG6857 and CG6881 both share an intron (same position and same phase) in TM3 (Table 3) with genes encoding both CCKR (accession #AF015959-AF015963) and human GASR (L10822). Likewise, all of these genes have an intron in a similar position within the highly variable cytoplasmic loop between TM5 and TM6. This conservation of introns further indicates that CG6857 and CG6881 are members of the CCKR/GASR receptor subgroup.
Locations and Phasing of Introns among Genes EncodingDrosophila Family A Peptide GPCRs
Family A/Group III-B: Neurokinin Receptors
The neurokinin (tachykinin) receptors (NKRs) are a monophyletic group of GPCRs that are also closely related to the orexin/hypocretin receptors (OXRs), the neuropeptide FF/AF receptor (NFFR), and a class of orphan, glucocorticoid-induced receptors (GCRCs). We found sixDrosophila members of this subgroup: CG5811 (Li et al. 1992b;St-Onge et al. 2000), CG10626, TAKR86C (Monnier et al. 1992), TAKR99D (Li et al. 1991, 1992b), CG10823, and BACR48G21.1. On the subgroup-specific tree (Fig. 1B), TAKR86C and TAKR99D were located near the base of a branch leading to NK1R-NK3R, which indicates that the two fly proteins (and the two C. elegans orthologs) arose before the diversification the vertebrate NKRs. TAKR86C and TAKR99D are located together on the subgroup tree (with stable fly neurokinin receptor, STKR; Fig. 1B), and in BLASTP searches, each detects the other with the lowest P values (Table 2). Moreover, the Takr86C (Rosay et al. 1995) and Takr99Dgenes share two introns in the same position and with the same phase (Table 3). Thus, TAKR86C and TAKR99D appear to be paralogs, and they are therefore likely to share similar ligands and functional properties.
Two additional Drosophila receptors, CG5811 and CG10626, are related to the true NKRs. However, based on BLASTP and phylogenetic analysis (Table 2, Fig. 1B), each of these receptors appears to be the ortholog of a NKR-related class of GPCRs that to date have been identified only in invertebrates. CG10626 is closely related to the tick NKR (LKR; Holmes et al. 2000) and the snail lymnokinin receptor (LSR) (Table 2), and these three receptors are located on a single branch of the subgroup-specific tree (Fig. 1B). Likewise, CG5811 displays strong sequence similarity with GRL106, a snail NKR-like protein (Table 2). The branching pattern of this portion of the NKR subgroup tree is unstable (Fig. 1B). However, CG5811 andCG10626 have two introns that are in similar locations and display the same phasing (Table 3). Thus, these genes appear to be paralogs that diverged independently of the true NKRs. Consistent with this interpretation, the midpoint root of the NKR subgroup tree is located between the branch leading to the true NKRs and the branches of the tree leading to CG5811, CG10626, and the related GPCRs.
Finally, there are two additional GPCRs, CG10823 and BACR48G21.1, that display moderate to weak homology with the NKRs. ByBLASTP, CG10823 displays strongest homology with the vertebrate neuropeptide FF/neuropeptide AF receptor (NFFR), the putative mammalian RF-amide-related peptide receptor (OT7T022; Hinuma et al. 2000), as well as CG5811 (Table 2). In addition, theCG10823 gene has an intron that is located in the same position (and phase) as one of the two introns shared byTakr86C and Takr99D (Table 3). On the subgroup-specific tree, CG10823 is located near the base of a branch leading to the orexin/hypocretin receptors (OXRs), OT7T022 and NFFR (Fig. 1B). Therefore, CG10823 appears to have arisen from a common ancestor of these vertebrate relatives. Finally, BACR48G21.1 also appears to be a member of the NKR subgroup. However, this relationship was not well supported by the phylogenetic analysis (Table 2), and additional sequence data will be required to evaluate this finding.
Family A/Group III-B: Neuropeptide Y Receptors
The receptors for the neuropeptide Y (NPY) family of peptides (NPYRs) and the prolactin releasing peptide (PRPR) form a subgroup of related GPCRs (Hinuma et al. 1998; Hoyle 1999). FourDrosophila proteins, CG1147, CG7395, CG12610, and CG13995, appear to be members of this subgroup (Table 2). On the subgroup-specific tree, the position of the root was unclear (Fig. 1C). In addition, although the branching pattern for the portions of the tree containing the vertebrate NPYR receptors (except NY2R) was stable, the rest of the tree was not clearly resolved. In theBLASTP analysis and on the phylogenetic trees (Table 2; Fig. 1C), CG1147 showed the strongest sequence homology with a class of receptors that includes a C. elegans orphan GPCR (C25G6.5) and the vertebrate neuropeptide Y Y2 receptors (NY2Rs). CG7395, which also displays strong general sequence homology with the other members of this subgroup, appears to be most closely related to a diversified group of orphan NPYR-like C. elegans receptors. In contrast, CG12610 appears to be most closely related to PRPR. The fourthDrosophila receptor in this group, CG13995, was located on the Group III portion of the full Family A tree, which consists almost exclusively of peptide GPCRs. However, CG13995 did not show strong evidence of homology with any specific class of peptide GPCRs (Table2). Interestingly, the CG13995 gene shares an intron in TM3 (same position and phase) with CG12610. Therefore, we propose that CG13995 is distantly related to the NPYR subgroup. Finally, it has been suggested that CG5811 is a NPYR-like receptor, despite its greater sequence similarity with the NKRs (see above), based on the activation of functionally expressed CG5811 by NPY and related peptides (at micromolar concentrations) and the lack of activation by vertebrate neurokinins (Li et al. 1992b). However, in competitive displacement experiments with CG5811 (St-Onge et al. 2000), PQGRF-amide-like peptides (e.g., NPFF and Lymnaea cardioexcitatory peptide) displayed IC50s in the subnanomolar range. Thus, CG5811 does not appear to be a member of the NPYR subgroup.
Family A/Group III-B: Bombesin/Gastrin Releasing Peptide Receptors
The bombesin-like neuropeptides, which include bombesin, gastrin releasing peptide (GRP), and neuromedin B (NMB), exert a wide variety of physiological actions in the CNS and the periphery through a class of related receptors (Sun et al. 2000). These receptors include the GRP-preferring receptor (GRPR), the neuromedin B-preferring receptor (NMBR), and an orphan class of receptors, characterized by bombesin receptor subtype 3 (BRS3). There are two Drosophila GPCRs, CG14484 and CG14593, that belong to this phylogenetic subgroup (Table2). On the subgroup-specific tree, the three types of vertebrate bombesin/GRP receptors formed a clade, whereas CG14484 and CG14593 branch out from the base of the tree (Fig. 1D). Therefore, it appears that the fly receptors diverged from a common ancestor of the vertebrate bombesin/GRP receptor lineages. The organizations of theCG14484 and CG14593 genes are similar; each has one intron in the same position and phase, and there are two additional introns in similar positions (Table 3). Thus, CG14484 and CG14593 appear to be paralogs. Together, these results indicate that CG14484 and CG14593 are bombesin/GRP receptors; to our knowledge, this is the first clear molecular evidence for bombesin/GRP signaling in invertebrates.
Family A/Group III-B: Growth Hormone Secretagogue, Neurotensin, Neuromedin U, and Thyrotropin Releasing Hormone Receptors
The receptors for neurotensin (NTR), neuromedin U (NMUR), thyrotropin releasing hormone (TRFR), and growth hormone secretagogue (GHSR) form a large and diverse subgroup of GPCRs (Fujii et al. 2000). Among these, NTR, GHSR, and NMUR display strong sequence similarity, whereas TRFR is more distantly related. At least sevenDrosophila GPCRs appear to be members of this subgroup: CG8784, CG8795, CG9918, CG14575, CG5911A, CG5911B, and CG14003 (Table2). An additional seven GPCRs (CG2114, CG5936, CG6986, CG8985, CG13229, CG13803, and CG16726) are all most closely related to a large set of related orphan receptors that had been identified previously only inC. elegans (C. Bargmann, pers. comm.). These orphan GPCRs fall into at least three classes, and there are one to threeDrosophila GPCRs in each class (Fig. 1E). The three receptors in class A (CG8985, CG13229, and CG13803) display strong sequence homology. In addition, CG8985 and CG13803 are linked genes (∼30 kb apart), and they share an intron (Table 3). Thus, theDrosophila class A receptors appear to be paralogs. All three classes display weak sequence similarity with TRFR, NTR, and GHSR, indicating that this entire family of orphan receptors may be derived from an ancestor of these vertebrate receptors and therefore may encode peptide GPCRs. However, confirmation of such a relationship will require functional analysis of one or more members of these orphan GPCR classes.
CG8784 and CG8795 are two of the seven Drosophila GPCRs displaying the strongest sequence similarity with this vertebrate subgroup, and they appear to be paralogs. They display strong sequence similarity with each other (Table 2; Fig. 1E). Moreover, theCG8784 and CG8795 genes are closely linked (∼10 kb apart) and share four introns with identical positions and phasing (Table 3). Similarly, CG9918 and CG14575 each share one intron with CG8784/CG8795 (Table 3), indicating that all four of these receptors are closely related. Their closest vertebrate homologs are NMUR, GHSR, and NTR, based onBLASTP analysis and on their positions in the phylogenetic trees (Table 2; Fig. 1E). Consistent with this finding, the shared intron located in the TM6 domain of CG8784 and CG8795 is also found in the same position and with the same phasing in the pufferfish GHSR gene (AF082211). However, the branching pattern for the subgroup-specific tree was unstable, and the evolutionary relationships among these receptors are unclear. Three additional receptors, CG5911A and CG5911B (generated by putative alternative splicing of the CG5911 gene) and CG14003, also displayed moderate to weak sequence homology with this subgroup and appear to be most closely related to vertebrate TRFR.
Family A/Group V: Galanin/Allatostatin and Opioid/Somatostatin Receptors
There were four Drosophila receptors, AlstR(Birgül et al. 1999; Lenz et al. 2000a), CG7285, CG10001 (Lenz et al. 2000b), and CG13702, that displayed strong sequence similarity with galanin, somatostatin, and opioid receptors (Table 2). Because these three classes of vertebrate receptors display extensive sequence similarity, we grouped them together to construct a subgroup-specific tree (Fig. 2A). The root of this tree is located between the branch leading to the galanin receptors and the branch leading to the somatostatin and opioid receptors. CG7285 and CG13702 were located on the branch containing all of the somatostatin and opioid receptors and related orphan GPCRs. The opioid receptors form a clade, and two groups of somatostatin receptors also form clades (SSR1/4 in one and SSR2/3/5 in the other). The remaining branches on this side of the tree are unstable. Together, these results indicate that CG7285 and CG13702 are orthologous to the vertebrate somatostatin and opioid receptors, although it is not clear whether they diverged from a common ancestor or from a point deeper within the tree. CG7285 and CG13702 appear to be paralogs; they display strong sequence homology (Table 2; Fig. 2A), and they are encoded by linked genes (∼90 kb apart) that share an intron with the same location and phasing (Table 3).
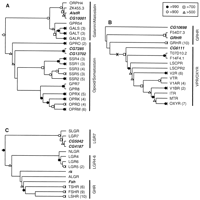
Neighbor-joining phylogenetic trees for the Family A, Group V receptors. (A) Rooted tree for the opioid, somatostatin, galanin, and allatostatin receptors. (B) Unrooted tree for the gonadotropin releasing hormone (GnRH), vasopressin, and oxytocin receptors. The likely midpoint of the tree is indicated with an “X.” (C) Rooted tree for the glycoprotein hormone receptors and related leucine-rich repeat containing receptors (LGRs). Bootstrap scores, omitted branches, and Drosophila GPCRs are indicated as in Fig. 1. (ALGR) Anthopleura elegantissima (sea anemone) LGR; (FSHR) follicle-stimulating hormone receptor; (GALR) galanin receptor type 1; (GALS) galanin receptor type 2; (GALT) galanin receptor type 3; (GPR24 and GPR54) mammalian orphan GPCRs; (GRHR) GnRH receptor; (ITR) isotocin receptor; (LGR4–7) LGR types 4–7; (LSCPR and LSCPR2) Lymnaea stagnalis conopressin receptor types 1 and 2; (LSHR) lutropin-choriogonadotropic hormone receptor; (MTR) mesotocin receptor; (NLGR) C. elegans LGR; (ORPH4) Lymnaea stagnalis orphan GPCR; (OPRD) delta-type opioid receptor; (OPRK) kappa-type opioid receptor; (OPRM) mu-type opioid receptor; (OPRX) nociceptin/orphanin FQ receptor; (OXYR) oxytocin receptor; (SLGR)L. stagnalis GRL101; (SSR1–SSR5) somatostatin receptor types 1–5; (TSHR) thyrotropin receptor; (V1AR and V1BR) vasopressin V1A and V1B receptors; (V2R) vasopressin V2 receptor; (VTR) vasostocin receptor. The remaining non-Drosophila sequences are orphan GPCRs from C. elegans.
The allatostatin receptor, AlstR (Birgül et al. 1999), and CG10001 were located on the portion of the tree containing all of the galanin receptors (Fig. 2A), indicating that AlstR and CG10001 are Drosophila orthologs of the mammalian galanin receptors. This finding is in agreement with an earlier phylogenetic analysis of AlstR (Birgül et al. 1999). The AlstR and CG10001genes share an intron at the same location and with the same phasing (Table 3; Lenz et al. 2000b). Thus, AlstR and CG10001appear to be paralogs and are likely to share many functional properties. Interestingly, immunocytochemical studies, using anti-porcine galanin and anti-porcine galanin message-associated peptide, as well as receptor autoradiography studies using125I-porcine galanin, showed the presence of galanin-like peptides in several locations in the adult CNS of blowflies, including the fan-shaped body of the central complex and a ring of cells in the medulla (Lundquist et al. 1991, 1993; Johard et al. 1992). Similar patterns of staining in the fan-shaped body and medulla have been obtained in Drosophila with a specific monoclonal anti-allatostatin antiserum (Yoon and Stay 1995). These comparative data provide additional support for the conclusion thatAlstR and CG10001 are closely related to the vertebrate galanin receptors (cf., Birgül et al. 1999; Lenz et al. 2000b).
Family A/Group V: Gonadotropin Releasing Hormone, Vasopressin, and Oxytocin Receptors
The receptors for gonadotropin releasing hormone (GRHR) and the receptors for vasopressin (VPR) and oxytocin (OXYR) belong to two closely related clades of GPCRs (Hoyle 1999). In Drosophila, there are three GPCRs that belong to this subgroup; CG6111, CG10698, and Dm-GRHR (Table 2; Hauser et al. 1998). The branching pattern near the base of the subgroup-specific tree was unstable (Fig. 2B), and the evolutionary history of this subgroup is unclear. However, when the tree is midpoint rooted, Dm-GRHR and CG10698 branch from the side of the tree leading to the vertebrate GRHRs, and CG6111 branches from the side of the tree leading to VPR, OXYR, and related GPCRs. These results are in agreement with the results of BLASTP analysis. Moreover, the Dm-GRHR gene shares an intron near TM4 (identical location and phasing) with the rat GRHR gene (U92471)(Hauser et al. 1998); CG10698 also shares this intron. Thus, Drosophila appears to have two GRHR-like receptors and one VPR/OXYR-like receptor.
Family A/Group V (Type 1c): Glycoprotein Hormone Receptors
Four glycoprotein hormones have been identified in mammals: thyroid-stimulating hormone (TSH) and the gonadotropins, follicle-stimulating hormone (FSH), choriogonadotropin (CG), and luteinizing hormone (LH). These four hormones bind to a subgroup of receptors (the LGRs) that all bear a characteristic, large, N-terminal “ectodomain” that participates in the binding of the large glycoprotein ligands (Hsu et al. 2000) (type 1c receptors; Bockaert and Pin 1999). Four Drosophila receptors, CG4187, CG5042, and the proteins encoded by the Fsh (Hauser et al. 1997) andrk (Ashburner et al. 1999; Eriksen et al. 2000) genes, display sequence similarity with the LGRs, including the N-terminal ectodomain (Table 2). On the subgroup-specific tree (Fig. 2C), there were three distinct clades (cf., Hsu et al. 2000). The first includes LGR7, aLymnaea ortholog (SLGR), CG4187, and CG5042. The second includes LGR4–LGR6, and the third includes the glycoprotein hormone receptors (LSHR, FSHR, and TSHR). Fsh is located at the base of a branch leading to the glycoprotein hormone receptors, indicating that this gene may have evolved from a common ancestor of LSHR, FSHR, and TSHR. Three additional receptors, C. elegans LGR (NLGR), sea anemone LGR (ALGR), and rk, were grouped only weakly with the glycoprotein hormone receptors; the branching pattern of this portion of the tree was unstable. Therefore, these could not be assigned to any one class of LGRs by basis of the phylogenetic analysis alone.
Within the ectodomain, all of the LGRs contain a variable number of leucine-rich repeats and a functionally important hinge region located between the leucine-rich repeats and the seven-TM core. At the borders of the hinge region, there are two sequences that are diagnostic of the three different subclasses of LGRs (Table4; Hsu et al. 2000). These groupings are also supported by BLASTP analysis of the ectodomains (data not shown). These sequences support the placement ofFsh in the subfamily of glycoprotein hormone receptors.
Conserved LGR Hinge Sequences
Placement of CG4187 and CG5042 in the LGR7 clade is supported byBLASTP analysis of the ectodomains (data not shown) and the presence of subgroup-specific hinge sequences (Table 4). Unlike the other two subgroups of LGRs, the ectodomains of LGR7 and snail LGR have low density lipoprotein (LDL) receptor-like cysteine-rich motifs at the N terminus (Tensen et al. 1994; Hsu et al. 2000). CG4187 and CG5042 also each contain at least one LDL motif (Table 4). The function of the LDL motif is unclear, but it indicates a possible role for lipoprotein-like molecules in neuronal G protein-mediated signal transduction (Tensen et al. 1994). Alternatively, given the presence of leucine-rich repeats, these receptors may bind to glycoproteins.
Although phylogenetic analysis of the LGRs did not place rk in any of the three subgroups of LGRs, analysis of the ectodomain indicates that this receptor is orthologous to LGR4–6. This is based on the presence of hinge sequences most similar to LGR4–6 and onBLASTP analysis (data not shown). The other members of this family are orphan receptors. However, the presence of the leucine-rich repeats indicates that these proteins also bind to glycoproteins.
Family B/Group I: Calcitonin and Diuretic Hormone Receptors
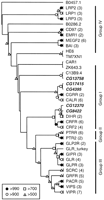
In addition to the 40 proteins in Family A (the rhodopsin-like receptors), there are 5 Drosophila peptide GPCRs in Family B (the secretin-like receptors). Based on BLASTP analysis and their positions on the phylogenetic tree (Fig.3), at least four of these receptors (CG4395, CG8422, CG12370, and CG17415) belong to Group I. This group contains the receptors for calcitonin (CALR), calcitonin gene related peptide (CGRR), corticotropin releasing factor (CRFR and CRF2), and diuretic hormone (DIHR). The position of the fifth Drosophila peptide GPCR in this family (CG13758) is unclear, and it may be a member of Group I, II, or III. CG8422 and CG12370 appear to paralogs, and they are orthologous to the DIHRs. These receptors belong to a clade containing CRFR and CRF2, which indicates that the ancestor to the insect DIHRs evolved from a common ancestor of the vertebrate corticotropin releasing factor receptors (Fig. 3). In contrast, CG4395 and CG17415 are most closely related to CALR and CGRR, although the bootstrap scores more deeply located within this branch of the tree were not strong enough to determine whether CALR and CGRR diverged before or after the related Drosophila receptors. We did not find evidence for well defined GPCR-associated proteins (e.g., RAMPs [Bockaert and Pin 1999] and RCPs [Evans et al. 2000]).
Unrooted neighbor-joining tree for the Family B receptors. The location of the tree midpoint is ambiguous and is therefore not indicated. Bootstrap scores, omitted branches, and Drosophila GPCRs are indicated as in Fig. 1. The four groups of Family B receptors are indicated with vertical bars. (BAI) brain-specific angiogenesis inhibitors 1–3; (CALR) calcitonin receptor; (CAR1) cyclic AMP receptor 1; (CD97) leucocyte antigen CD97; (CGRR) calcitonin gene-related peptide type 1 receptor; (CRF2) corticotropin releasing factor (CRF) receptor 2; (CRFR) CRF receptor 1; (DIHR) diuretic hormone receptor; (EMR1) cell surface glycoprotein EMR1; (GIPR) gastric inhibitory polypeptide receptor; (GLP2R) glucagon-like peptide 2 receptor; (GLPR) glucagon-like peptide 1 receptor; (GLR) glucagon receptor; (GRFR) growth hormone releasing hormone receptor; (HE6) G protein-coupled receptor HE6; (LRP1–3) calcium-independent alpha-latrotoxin receptors (latrophilins) 1–3; (MEGF2) seven-pass transmembrane proteins CELSR1–2 and MEGF2; (PACR) pituitary adenylate cyclase activating polypeptide (PACAP) type I receptor; (PTR2) parathyroid hormone receptor; (PTRR) parathyroid hormone/parathyroid hormone-related peptide receptor; (SCRC) secretin receptor; (TM7XM1) human EGF-TM7 like protein; (VIPR) vasoactive intestinal polypeptide (VIP) receptor 1; (VIPS) VIP receptor 2. The remaining non-Drosophila sequences are orphan GPCRs from Caenorhaloditis elegans.
Drosophila Genes Encoding Neuropeptides and Peptide Hormones
We wished to compare the number of peptide GPCRs with the number of neuropeptides present (or suspected to exist) in Drosophila. Based on the literature and on some genomic analysis, we have assembled a list of 22 Drosophila neuropeptide genes (Table5). These genes are either known or predicted to encode bioactive neuropeptides and peptide hormones. Eight of these, which encode neuropeptides described for Drosophilaor other arthropods, were described previously only in gene annotations generated by Celera/BDGP and in a parallel survey, which was just published recently (Vanden Broeck 2001). An additional peptide listed by Vanden Broeck (2001) (“IFa”) was not included, because the precursor did not match our criteria for putative neuropeptide genes. Because neuropeptide-encoding precursors do not display multiple, uniform characteristics found in GPCRs, we are certain to have missed many peptide genes and thus consider this list incomplete. However, assuming a 1 : 1 ratio of neuropeptide and peptide hormone genes to peptide GPCRs, these 22 genes appear to encode the ligands for at least 50% of the Drosophila peptide GPCRs that we have described. This may be an underestimate, given the fact that many of these neuropeptide genes encode multiple peptides. In addition to these 22 neuropeptide genes, we list several insect peptides and peptide hormones known in other insects and for which Drosophilahomologs have been inferred by observation or simply by conjecture. Although the structures of these genes are not yet available, they are included here to permit consideration of all plausible ligands for the identified peptide GPCRs.
Drosophila Neuropeptides and Peptide Hormones
Ligands for Family A Peptide GPCRs
There are multiple genes encoding potential ligands for theDrosophila NKR-like receptors. CG14734 produces neurokinin-like peptides (Siviter et al. 2000) that likely bind to TAKR86C and TAKR99D, as shown by functional expression of these receptors and specific binding to mammalian (Li et al. 1991) and insect (Monnier et al. 1992) neurokinins and related peptides. Based on the pharmacological observations of the Lymnaea lymnokinin receptor, LSR (Cox et al. 1997), we speculate that theDrosophila ortholog, CG10626, is a receptor for the leucokinin-like peptides encoded by CG13480 (Terhzaz et al. 1999).
In addition to the neurokinin-like peptides, there are also several genes that are known to encode (or potentially encode) peptides terminating in the sequence RF-amide. These include the putative peptide products of a novel gene, CG13968. Along with the products of the dFMRFa (Nambu et al. 1988; Schneider and Taghert 1988) andDMS (CG6440) genes, these peptides may interact with multiple receptors for RF-amide peptides. CG5811 has been shown to bind with high affinity to molluscan -PQGRF-amide peptides and therefore is likely to represent the first of several Drosophila RF-amide peptide receptors (St-Onge et al. 2000). A second potential RF-amide peptide receptor, CG10823, is orthologous to NFFR and OT7T022, both of which bind ligands bearing the C-terminal consensus sequence PXRF-amide (Elshourbagy et al. 2000; Hinuma et al. 2000). The otherDrosophila neuropeptides ending in RF-amide are found within the dsk gene and display structural similarity to the vertebrate cholecystokinins (Nichols et al. 1988). We speculate that these peptides may interact with either or both of the paralogous CCKR/GASR-related receptors (CG6857 and CG6881).
AlstR binds a native Drosophila allatostatin peptide (AST-1) with high affinity (Birgül et al. 1999). This peptide, along with multiple other related peptides, is encoded by CG13633 (Lenz et al. 2000c). Because AlstR and CG10001 are paralogs, the latter receptor is also likely to interact with one or more of the products ofCG13633.
Ligands for Family B Peptide GPCRs
We speculate that the corticotropin releasing factor (CRF)-related peptides encoded by CG8348 and CG13094 (similar to Locustadiuretic hormone; Coast 1996; Furuya et al. 2000) interact with the CRFR-related CG8422 and/or CG12370, both of which are orthologs (Fig.3) of the Acheta domesticus diuretic hormone receptor (Reagan 1994). Additionally, Zhong and Pena (1995) found evidence for a PACAP-like peptide in flies. Feany and Quinn (1995) and Moore et al. (1998) provided genetic evidence to implicate the amnesiacgene (potentially encoding peptides of the PACAP family) in variousDrosophila behaviors. We speculate that the peptides in this group may interact with one or more of the remaining Family B receptors (CG4395, CG17415, and CG13758).
Peptide Genes Still Awaiting Identification
There are several insect neuropeptides and peptide hormones that have not as yet been cloned in Drosophila. These include three large protein hormones—PTTH, bursicon, and the anterior retraction factor (ARF)—that are known to exist in Drosophila but currently lack molecular definition. At least two of these proteins, PTTH and bursicon, are glycoprotein hormones (Fraenkel et al. 1966; Kim et al. 1997), whereas the structure of ARF remains undefined (Sivasubramanian et al. 1974). As noted above, the structure of the receptor encoded by the fsh gene indicates that it binds to a glycoprotein hormone ligand. All of the mammalian glycoprotein hormones share a similar structure, consisting of common α- and specific β-subunits (Hsu et al. 2000). However, to date, no similar proteins have been identified in flies. We speculate that PTTH, bursicon, and ARF are good candidate ligands for members of the LGR class of receptors.
Relatives of several peptides identified in other insects may also be present in Drosophila. These include PBAN and diapause hormone (DH), which are found in diverse insects. Both are peptide hormones of moderate size that are cosynthesized along with shorter pyrokinin peptides (Kawano et al. 1992; Sato et al. 1993; Masler et al. 1994; Xu et al. 1995). It is notable that the Drosophila CG15520 precursor includes a single FXPRLamide (pyrokinin-like) peptide but lacks any sequences similar to PBAN or DH. Because NMUR is activated by peptides displaying a LXXPRX-amide consensus (Fujii et al. 2000), we speculate that this pyrokinin-like peptide may interact with CG14484 and/or CG14593, which are orthologs of NMUR. Likewise, theDrosophila ecdysis triggering hormones (ETHs), which have a PRX-amide C-terminal sequence (Park et al. 1999), may signal through these receptors.
With the completion of the D. melanogaster genome sequence, we are now able to take a comprehensive picture of the genes encoding peptide GPCRs in this species, and a complete catalog of the cognate ligands should soon follow. This is an important first step toward detailed physiological and genetic analyses of neuropeptide signaling in Drosophila.
METHODS
Peptide GPCR Sequence Acquisition
To identify all predicted Drosophila GPCRs, we first scanned the gene annotations developed jointly by the BerkeleyDrosophila Genome Project (BDGP) and Celera Genomics for all proteins predicted to contains domains matching seven-TM motifs (Adams et al. 2000; Brody and Cravchik 2000). We rejected sequences identified recently as odorant receptors by a committee representing scientists working in the field ( Drosophila Odorant Receptor Nomenclature Committee 2000). Each remaining cloned and candidate receptor gene was used as a BLASTP search query of the database of predicted Drosophila proteins using the BDGP server (http://www.fruitfly.org/) and/or of the “non-redundant” database of all proteins using the NCBI server (http://www.ncbi.nlm.nih.gov/). Sequences were not considered further if the resulting top-scoring proteins yielded P values for nonpeptide receptors (and associated orphan receptors) that were at least 10-fold greater than the top P value for a putative peptide receptor. Three sequences (CG18314, CG12796, CG13579) generated a smaller range of P values following BLASTPsearches on the BDGP server. Nevertheless, analysis of these proteins using the NCBI server yielded hits that were exclusively amine/small neurotransmitter receptors (or orphan receptors). These proteins therefore are likely to encode nonpeptide receptors, and they also were excluded.
We first scanned a set of GPCR sequence annotations obtained through aGENSCAN search of the complete Drosophila genome sequence (see Vosshall et al. 1999) and identified based on sequence similarity to GPCRs in the NCBI nonredundant protein database (K. Scott and L. Vosshall, pers. comm.). For the BLASTP andTBLASTN analyses, we assembled a “GPCR query set,” which included the previously annotated peptide, amine, and related/unclassified Drosophila GPCRs as well as ∼200 sequences representing a diverse set of Family A (rhodopsin receptor-like family) GPCRs from the Pfam database (7TM-1; http://pfam.wustl.edu/). These sequences were used as queries for BLASTP andTBLASTN searches on the BDGP server, using the predicted proteins and the Celera/BDGP whole genome shotgun sequence datasets, respectively. To expedite the latter search, we assumed thatTBLASTN hits to genomic sequences that were already on our list were due to the detection of previously annotated GPCR genes.
GPCR Alignments
We used the hidden Markov model–based protein alignments contained in Version 5.5 of PFAM (Sept., 2000; Bateman et al. 2000) as a template for the manual alignment of the Drosophilacloned and candidate peptide receptors. The alignments were viewed using ClustalX (Version 1.8; Thompson et al. 1997), and, in some cases, this program was used to help resolve the alignment of variable regions (e.g., between TM domains 4 and 5). We used these alignments to build phylogenetic trees and also to detect missing or incorrect sequences in the gene annotations.
The N-terminal and C-terminal non-TM sequences in GPCRs tend to be poorly conserved, making accurate alignment difficult, and the seven-TM core region is sufficient for the subclassification of these proteins (Strader et al. 1994). Therefore, for Family A receptors, we deleted sequences N-terminal to the conserved GNXXLV motif (single-letter amino acid code) in TM1 and C-terminal to the conserved NPXIY motif in TM7. For Family B receptors (secretin receptor family), we deleted sequences flanking the X10GX3S motif in TM1 and the QGX2V X4CX5X motif in TM7.
Correction of Annotations
To locate missing TM domains among the putative peptide receptor annotations, we scanned for potential coding exons in flanking genomic sequence using the GENSCAN server at MIT (http://genes.mit.edu/GENSCAN.html), and the FGENES (gene prediction) and FEX (exon prediction) programs on the Baylor College of Medicine (BCM Search Launcher) server (http://www.hgsc.bcm.tmc.edu/). We also scanned for potential mRNA splice sites using the SPL program on the BCM Search Launcher server and by manual inspection of potential open reading frames displayed using MacVector (Genetics Computer Group, Madison, WI). The DNA sequences for all of the predicted donor and acceptor splice sites were NN‖GT and AG‖NN, respectively. Finally, we examined neighboring gene annotations to identify duplicate annotations of single GPCR genes. The annotations were judged to be complete when each of the TM domains displayed features that were clearly recognizable among closely related receptors. Except for the LGR subgroup of receptors (see Results and Discussion), which all share a large and subgroup-specific N-terminal domain, we did not evaluate the quality of the annotations for the N-terminal and C-terminal non-TM regions.
Tree Building
We classified the cloned and candidate peptide GPCRs based on five criteria. First, we noted the highest scoring BLASTP hits obtained on the NCBI server (Table 2). Second, we constructed alignments of Family A and Family B receptors, including all of theDrosophila peptide GPCRs identified above, for the purpose of generating full phylogenetic trees for each family. For Family A, we included mostly complete (TM1–TM7) sequences representing each of the five receptor groups, as well as sequences representing each of the various subgroups of receptors (e.g., the three types of galanin receptors) and representative orphan receptors contained within the full list of Pfam 7TM-1 (Family A) GPCRs. For Family B, we included all of the Group I–III receptors and a representative set of Family B, Group IV receptors within the full list of Pfam 7TM-2 GPCRs. After manual editing of the alignments, we constructed neighbor-joining phylogenetic trees for each family using ClustalX, using the correction for multiple substitutions provided by the software, followed by bootstrap analysis (1000 replicates).
For the subsequent subgroup-specific trees, we attempted to include all complete TM1–TM7 sequences belonging to each subgroup (as well as some partial sequences). These were identified by scanning the full Pfam 7TM-1 alignment and the GPCRDB listing of available GPCR sequences (http://www.gpcr.org/7tm/), and by performing BLASTPsearches with the cloned and candidate Drosophila peptide GPCRs as well as other representatives of each subgroup. After manual editing of the alignments, the construction of neighbor-joining trees and the bootstrap analysis was performed as above. A set of 26 indoleamine (biogenic amine) receptors, which form a monophyletic group (Kolakowski 1994), was used as an outgroup for the purpose of rooting the subgroup-specific trees. When the position of the root was unclear, the outgroup was omitted. All alignments, revised annotations, and unabridged versions of the trees are located athttp://thalamus.wustl.edu/flyGPCR/peptideGPCR.html. In addition, the revised annotations have been submitted to FlyBase (http://flybase.bio.indiana.edu/).
Acknowledgments
This work was supported by National Institutes of Health Grant NS21749 and the Human Frontier Science Program Organization (P.H.T.). We thank Sean Eddy for helpful discussions, Kirstin Scott and Leslie Vosshall for sharing Drosophila GPCR sequence data, Lin Yang and Dori Sztipanovits for technical assistance, and Aguan Wei for comments on the manuscript. We also thank Cori Bargmann and Kemal Payza for sharing unpublished results.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
We have identified two additional ESTs for peptide GPCRs: AT008361(CG1147) and AT0019640 (CG13229). Also, based on discussions with Jan Veenstra (Universite Bordeaux) we now add two additional peptide genes to our list: the SIFamide gene (currently listed as part ofCG4681; Ifa, Vanden Broeck 2001) and the hugingene (CG6371).