Alpha Satellite DNA Variant-Specific Oligoprobes Differing by a Single Base Can Distinguish Chromosome 15 Homologs
Abstract
The ability to distinguish homologous chromosomes is a powerful cytogenetic tool. However, traditional techniques can only distinguish extreme physical variants and are highly dependent on sample preparation. We have previously reported oligonucleotide probes, specific for human chromosome 17 alpha satellite DNA sequence variants, that distinguish cytogenetically normal homologous chromosomes by FISH. Here we report the development of similar oligoprobes, differing at a single nucleotide position, that not only distinguish homologous chromosomes 15 but can be used to follow the transmission of a chromosome from parents to their offspring. We also identified a novel array-size polymorphism in another family. The alphoid array of one chromosome is quite small and below the detection threshold for our oligoprobes, although it is detectable by conventional FISH probes. This size polymorphism provides an additional FISH-based method for distinguishing homologs. Most importantly, this work illustrates the potential applicability of the technique to the entire human chromosome complement.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF234768, AF234769, AF234770,AF234771, AF234772, AF2234773, AF234774, AF234775, AF234776, AF234777, and AF237720.]
Chromosome heteromorphisms, plentiful in the human population, have been widely used not only to distinguish one human chromosome pair from another but also to distinguish chromosome homologs. Such visible physical differences between homologs were used to map the very first gene to a human autosome, chromosome 1 (Donahue et al. 1968). These heteromorphisms are common, stable, heritable, and are rarely affected by crossing over (Jacobs 1977). However, only extreme variants, which are rare within the population, can be reliably scored (Sherman et al. 1991; Lorber et al. 1992). This is partially attributable to the fact that the structural morphology of such variants can change depending on the chromosome preparation or banding technique used.
Centromeric heterochromatin, once considered “junk DNA,” is a rich and underutilized source of polymorphisms within the human genome (O'Keefe et al. 1996). Numerous studies have revealed the existence of sequence polymorphisms within the alpha satellite DNA arrays of human chromosomes. Alpha satellite DNA is a tandemly repeated DNA family that resides at the centromeres of all primate chromosomes. It is based on a 171-base-pair monomer that is further organized into higher-order repeat units (Manuelidis and Wu 1978; Willard 1985; Waye et al. 1987). Chromosome-specific subfamilies of alpha satellite DNA, which differ in the number of monomers within a higher-order repeat as well as in the restriction enzymes used to define the repeat, have been identified. These chromosome-specific subsets of alpha satellite DNA are also defined by sequence differences (Willard 1985; Jorgensen et al. 1986). Sequence analysis of several chromosomes has also identified polymorphisms within and between the alphoid arrays of individual chromosomes, although very few chromosomes have been analyzed in detail (Durfy and Willard 1989; Warburton and Willard 1995). Because sequence differences between homologs are entirely independent of preparation techniques, these variants can be exploited to reliably distinguish cytogenetically indistinguishable homologous chromosomes.
Previously (O'Keefe et al. 1996), we reported the development of oligonucleotide probes specific for sequence variants within the alpha satellite DNA array of human chromosome 17 for use in fluorescence in situ hybridization (FISH). A moderate amount of sequence data is required to effectively design alpha satellite variant-specific oligoprobes. Unfortunately, a very limited number of studies have investigated sequence variations within the centromere of a single chromosome.
Oligonucleotide probes also have the ability to distinguish single-base sequence differences (Conner et al. 1983). Such variant-specific oligoprobes have potentially powerful applications in both basic research and clinical laboratories. As an example, we have used these probes to study the behavior of chromosome 17 during normal male meiosis (O'Keefe et al. 1997). However, to realize this power, the technique must be extended to other human chromosomes and, ultimately, the entire human chromosome complement. The chromosomes for which such probes would be particularly useful are those that exhibit parent-of-origin effects in aneuploidy, imprinting, and cancer, including chromosomes 15, 16, and 21.
Here we report the development of oligonucleotide probes, specific for human chromosome 15, which distinguish cytogenetically normal homologous chromosomes by FISH. The probes hybridize to cloned sequences identical to those from which they were designed but not to single-base variants. In addition, these oligoprobes can be utilized to follow the transmission of a single chromosome from parent to offspring.
RESULTS
Genomic Organization and Sequencing of Chromosome 15 Alpha Satellite DNA
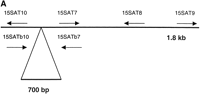
To design variant-specific oligonucleotide probes for chromosome 15, we first needed to sequence chromosome 15-specific alpha satellite DNA. Previous studies (Choo et al. 1990) had identified two independent subfamilies of chromosome 15-specific alphoid DNA. One subfamily, with a reported higher-order repeat of 2.5 kb, was identified by the clone pTRA-20. We sequenced the 1.8-kb insert of pTRA-20 in its entirety (GenBank accession no. AF27720) and used this sequence to develop chromosome 15-specific PCR primers (15SAT7, bases 458–479; 15SAT8, bases 1444–1423; 15SAT9, bases 1772–1793; 15SAT10, bases 372–354; see Fig. 1A). We performed PCR on DNA isolated from hybrid cell lines that contained a single human chromosome 15 to generate chromosome-specific products. To verify the specificity of our PCR products, we nick-translated the products and used them in a FISH assay on normal human metaphase preparations. The products hybridized exclusively to the centromeric region of chromosome 15 under high-stringency conditions (data not shown).
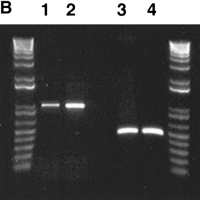
Genomic organization of pTRA-20-specific alpha satellite DNA. (A) A schematic of the 1.8-kb higher-order repeat of chromosome 15 along with the positions of the PCR primers (not drawn to scale). The primers were designed using pTRA-20 sequence. The location of the additional 700 bp is indicated below the 1.8-kb fragment. (B) PCR of monochromosomal somatic cell hybrids A9 + 15 (lanes 1 and 3) and GAR-1 (lanes 2 and4) with primer pairs 15SAT7/8 (lanes 1 and2) and 15SAT9/10 (lanes 3 and 4). 15SAT7/8 amplified a band of the predicted size, 986 bp. 15SAT9/10 amplified a band of 464 bp, indicating that the additional 700 bp is not adjacent to the 1.8-kb pTRA-20 sequence. (C) Southern blots of A9 + 15 and GAR-1 genomic DNA, digested with AccI (A), EcoRI (I), EcoRV (V), HincII (Hc), HindIII (Hd), and StuI (St). The top arrow marks 2.5 kb and the bottom arrow marks 1.8 kb. Both cell lines contain pTRA-20-specific higher-order repeats of 2.5 kb (EcoRV) and 1.8 kb (EcoRI).
We initially assumed that the pTRA-20 clone was a subfragment of the 2.5-kb higher-order repeat and that an additional 700 base pairs of sequence lay adjacent to the 1.8-kb pTRA-20 sequence. However, primers designed to amplify this smaller fragment (15SAT9 and 15SAT10; see Fig.1A) did not amplify a product of the predicted size when used on cell lines containing a single chromosome 15 (Fig. 1B). Primers designed to amplify sequences internal to the pTRA-20 sequence (15SATb7, bases 479–458; and 15SATb10, bases 354–372; see Fig. 1A) did produce products that contained the additional 700 bp from all cell lines (data not shown).
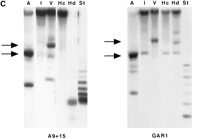
Southern blot analyses of the same hybrids support the conclusion that the alphoid subfamily identified by pTRA-20 has higher-order repeats of both 2.5 kb and 1.8 kb (Fig. 1C). pTRA-20 also hybridizes to high-molecular-weight DNA in many of the lanes shown. In principle, these bands might be due to cross-hybridization with other chromosome 15-specific repeats that lack these diagnostic restriction enzyme sites. However, this explanation is unlikely because we washed the filters at the same high stringency previously determined to distinguish pTRA-20-specific alpha satellite DNA from the other chromosome 15 alphoid DNAs (Choo et al. 1990). Because some enzymes (e.g., HindIII, StuI; see Fig. 1C) digest the high-molecular-weight fraction to completion, we favor the idea that the undigested material corresponds to the pTRA-20 family.
We and others have shown that the pTRA-20-specific arrays can differ greatly between individual chromosomes (Choo et al. 1990; this work). For example, HindIII reduces the A9 + 15 arrays to the 171-bp alpha satellite monomer, while GAR1 DNA is only partially digested into bands of 1.8 kb or greater (Fig. 1C). This variation in restriction digest patterns between the alphoid arrays of homologs has been observed for other alphoid subfamilies as well (e.g., Warburton and Willard 1995). The differences between chromosomes 15 are more pronounced in our studies than those of Choo et al. (1990); the differences are most likely due to the fact that we analyzed cell lines that are monochromosomal for chromosome 15 and not diploid cell lines. Overall, our Southern blot data illustrate that the organization of the pTRA-20-specific alpha satellite DNA is complex and most likely contains other differentially-sized higher-order repeats (Choo et al. 1990).
To obtain chromosome 15-specific alpha satellite DNA for sequencing, we used primers 15SAT7 and 15SAT8 to amplify a portion of the higher-order repeat. Due to the repetitive nature of alpha satellite DNA, primer annealing was performed at relatively high temperatures (60°C) to generate only the product of interest. At lower temperatures (58°C), multiple nonspecific bands were observed (data not shown). However, amplification at high melting temperatures greatly reduced the total amount of product. Therefore, to generate sufficient amounts of product, we performed PCR at 59°C and gel-purified the bands of interest. We then cycle-sequenced the purified PCR products at a high annealing temperature with one of the alphoid-specific primers, 15SAT7. This protocol allowed us to sample a large number of higher-order repeats from a single chromosome in one sequencing run.
Because we sequenced a heterogeneous pool of products, we were able to identify heteromorphic sites. For an individual sequence variant to be truly useful in distinguishing homologs, it must be present in a large number of the sequences. Ideally, a variant should be found in ∼40%–60% of the sequences within the pool.
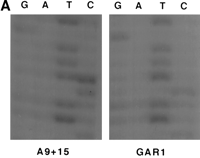
We sequenced PCR products from a total of seven hybrid cell lines over approximately 150 bases and found a single variant at position 529 (numbering corresponds to position in pTRA-20; Fig.2A). In the group of seven chromosomes studied, we identified two classes of chromosome 15. One class solely contained higher-order repeats with a T at position 529 (GAR1; Fig. 2A), while the second class contained higher-order repeats with either T or C at that position (A9 + 15; Fig. 2A). Thus, cycle-sequencing identified two distinct sequence variants of chromosome 15 alpha satellite DNA that differ at a single nucleotide position. Previous experience (Warburton and Willard 1992, 1995) predicts that a third class of chromosome 15 exists containing higher-order repeats that only exhibit a C at position 529. However, we did not identify such a chromosome during our sequencing efforts.
Sequence variation within chromosome 15-specific alpha satellite DNA. (A) Sequences of cell lines A9 + 15 and GAR1. Total PCR products, generated with primer pair 15SAT7/8, were sequenced using primer 15SAT7. A9 + 15 shows bands in the T and C lanes at position 529, while GAR1 shows a single band in the T lane. (B) Sequences of individual alpha satellite DNA clones. PCR products from both cell lines were cloned and sequenced individually. Clones from A9 + 15 have a T or a C at position 529, while clones from GAR1 only have a T.
To verify our findings, we TA cloned individual PCR products from the hybrid cell lines mentioned above. These clones were then sequenced using vector primers, yielding data consistent with our earlier results (Fig. 2B). Clones derived from cell lines that had either a T or a C at position 529 either had T or C, respectively, at the same position (A9 + 15 clones C1-C4, F1 and F2; GenBank accession numbersAF234768–234773). The clones derived from chromosomes with only a T at position 529 only had T, and no clones from these cell lines were found to contain a C (GAR1 clones E1, E2, E4 and E5; GenBank accession numbers AF234774–234777).
Oligonucleotide Probe Design and Specificity
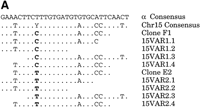
We used the higher-order repeat-variant sequences to design oligonucleotide probes specific to the two sequence variants of chromosome 15 alpha satellite DNA. Several versions of the oligonucleotide probes, differing in size and in the location of the discriminating base, were tested (see Methods). Of these probes, probe pair 15VAR1.4/15VAR2.4 was best able to distinguish homologs. Figure3A compares the sequences of the variant-specific probes (15VAR1.4 and 2.4, bases 524–546; Fig. 3A) with the overall and chromosome 15-specific alpha satellite DNA consensus sequences. The probes differ from the overall alphoid consensus at four or five positions, while they differ from each other solely at the variant position 529.
Design and specificity of variant-specific oligonucleotide probes. (A) The sequences of all variant-specific oligoprobes, including 15VAR1.4 and 15VAR2.4, are compared to the total and the chromosome 15-specific alphoid consensuses, as well as to clones E2 and F1. The variant position 529 is shown in bold. (B) The variant-specific probes in a Southern blot assay against clones E2 and F1. At low stringency, there is extensive cross hybridization. At high stringency, the probes are specific for the sequences from which they are designed.
To determine the specificity of 15VAR1.4 and 2.4, we tested them in a Southern blot assay against clones containing each variant (p15/78E2 and p15/78F1; Fig. 3A). Probe 15VAR1.4 was an exact match for clone p15/78F1 and differed from clone p15/78E2 at position 529. The opposite was true for 15VAR2.4. We found that at relatively high stringencies (hybridization and stringency washes in 30% formamide/2XSSC at 37°), 15VAR1.4 hybridized only to clone p15/78F1, while 15VAR2.4 was specific for p15/78E2 (Fig. 3B). Though the oligonucleotide probes differed at a single base, they distinguished chromosome 15 alpha satellite DNA sequence variants at high stringency.
Fluorescence In Situ Hybridization Studies
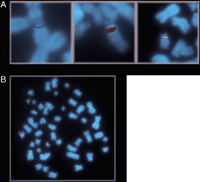
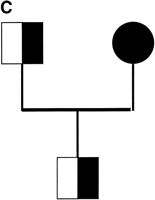
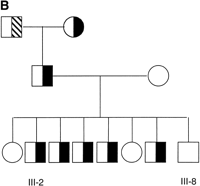
We used the variant oligoprobes in a dual-color FISH assay on metaphase chromosome preparations of CEPH cell lines. The haptenated variant-specific oligoprobes were hybridized and detected sequentially (see Methods). Upon scoring preparations from a number of CEPH individuals, we found all three predicted classes of chromosome 15: those positive for 15VAR1.4 only (green signal; left panel, Fig.4A), those positive for 15VAR2.4 only (red signal; center panel, Fig. 4A), and those positive for both oligoprobes (yellow signal; right panel, Fig. 4A). In our sequencing assay, three of the chromosomes had the T variant (red signal, see above) and four had the T/C variant (yellow signal). However, when we scored an additional 76 chromosomes by FISH with our variant-specific probes, we found that the majority of chromosomes screened, 64, were of the T/C variant class, four were of the T-only class, and eight were of the C-only class. When these probes were used to screen several pedigrees, we were able to identify a family in which one of the parents had distinguishable chromosomes 15 (Fig. 4B). Using our variant-specific oligonucleotide probes, we were able to follow the transmission of one chromosome 15 from parents to child (Fig. 4C).
Variant-specific oligonucleotide probes distinguish homologous chromosomes by FISH. (A) Three classes of chromosome 15 are identified by the oligoprobes: those positive for 15VAR1.4 only (green; left panel), those positive for 15VAR2.4 only (red; center panel), and those positive for both 15VAR1.4 and 15VAR2.4 (yellow; right panel). (B) 15VAR1.4 (signal in green) hybridizes to both chromosomes from this individual (father in pedigree from panel C), while 15VAR2.4 (signal in red) hybridizes only to one. (C) 15VAR1.4 and 15VAR2.4 were used to follow the transmission of a single chromosome from parents to offspring. Half-shaded symbols, 15VAR1.4/15VAR2.4 heterozygotes; shaded symbol, 15VAR2.4 homozygote.
Size Variation of Chromosome 15 Alpha Satellite DNA Arrays
During the course of our oligoprobe studies, we identified a family in which roughly half of the members displayed a detectable FISH signal on only one of the two chromosome 15. This finding was not due to failure of hybridization of the oligoprobes because the other homolog was positive for probe hybridization. In principle, the lack of signal on the homolog might be due to variant pTRA-20 sequences sufficiently different from the probe, preventing hybridization at the stringencies used. Alternatively, this chromosome may lack pTRA-20-specific alpha satellite DNA. Chromosome 15 contains at least two distinct subfamilies of alpha satellite DNA (Choo et al. 1990). The loss of the entire pTRA-20-specific array may not have any effect on the stability of a chromosome because another alpha satellite subfamily or other cryptic sequences may be able to form an active kinetochore. Although both chromosomes 15 in this family appeared to be normal on a gross cytogenetic level, we wanted to determine if both chromosomes contained the pTRA-20-specific subfamily of chromosome 15–specific alpha satellite DNA.
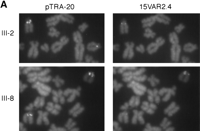
Hence, we nick-translated pTRA-20 and hybridized it to metaphase chromosome preparations from a number of family members. Those individuals that were positive for oligonucleotide hybridization on both chromosomes were positive for pTRA-20 on both chromosomes (see individual III-8; Fig. 5A). Interestingly, individuals that had only one chromosome 15 signal using oligoprobes were positive on both chromosomes when pTRA-20 was used, but one homolog had a much smaller pTRA-20 signal (individual III-2; Fig. 5B). We then hybridized pTRA-20 in the presence of one of the oligonucleotide probes (15VAR2.4) and found that the chromosome with the small pTRA-20 signal was the chromosome that was negative for oligonucleotide probe hybridization (Fig. 5B).
Size and oligoprobe signal variation of chromosome 15 alpha satellite DNA. (A) Partial metaphases from two individuals, hybridized with pTRA-20 and 15VAR2.4. Individual III-2 has one large and one small pTRA-20 signal. The chromosome with the large pTRA-20 signal is positive for 15VAR2.4, while the chromosome with the small pTRA-20 signal is negative for 15VAR2.4. Individual III-8 has two large pTRA-20 signals and is positive for 15VAR2.4 on both chromosomes. (B) Mendelian inheritance of a small pTRA-20 array and no oligonucleotide probe signal. Open symbol, 15VAR1.4–15VAR2.4 and large pTRA-20 array homozygote; half-cross hatched symbol, 15VAR1.4–15VAR2.4/15VAR2.4 heterozygote and large pTRA-20 array homozygote; half-shaded symbol, 15VAR1.4–15VAR2.4 hemizygote and large pTRA-20 array/small pTRA-20 array heterozygote.
Previously, Verma and Luke (1992) reported a family in which a chromosome 21 had a low alpha satellite DNA content and was undetectable by interphase FISH. More recently, Lo et al. (1999)further investigated several chromosomes including 13, 17, and 21 that had reduced centromeric signals and determined that these arrays were approximately 50–100 kb in size. The detection limit of our fluorescent oligoprobes has been estimated at approximately 100 copies (Matera and Ward 1992; C. O'Keefe, unpubl.). The pTRA-20-specific alpha satellite DNA subfamily has a higher-order repeat of up to 2.5 kb. In principle an alphoid array could be as large as 250 kb and not be visible by oligo-FISH. If the chromosome 15 alpha satellite DNA array on this chromosome is on the same order of size as the arrays studied by Lo et al. (1999), it would also not be detectable with our oligonucleotide probes. Though extremely small alpha satellite DNA arrays have been thus far reported for chromosomes 13, 17, and 21, we believe that the chromosome 15 in this family also contains an extremely small alphoid array and that its small size accounts for the lack of a signal visible to the oligonucleotide probes.
DISCUSSION
Here we report the development of oligonucleotide probes, specifically for sequence variants of chromosome 15 alpha satellite DNA, which can distinguish cytogenetically normal homologs by FISH. We further defined the organization of the pTRA-20 alphoid array of chromosome 15 and generated sequence information for this chromosome-specific alpha satellite DNA subfamily. Most chromosomes 15 scored using these oligonucleotide probes were of the same class, bearing both heteromorphic bases (529 T/C) within their alphoid arrays. However, we were able to identify three distinct classes of chromosome 15 and were able to follow the transmission of a single chromosome 15 from parent to child. The studies outlined above illustrate that alpha satellite DNA variant-specific oligoprobes can be designed for most, if not all, of the human chromosome complement.
Technical Considerations
Many issues important to the design of alpha satellite DNA variant-specific oligonucleotide probes arose during our studies of chromosome 15. Of greatest importance was the amount of sequence data available for an individual chromosome. Most studies on the sequence of chromosome-specific alpha satellite DNA families have used a small number of clones to generate a consensus higher-order repeat sequence. However, a high level of sequence variation between the arrays of homologs can be found when large numbers of clones are sequenced. We found a variant of chromosome 15 alpha satellite DNA within approximately 130 bases of sequence. Though time consuming, we believe that further efforts to sequence chromosome-specific alpha satellite DNA subfamilies will identify even more variants. A larger amount of sequence data will also allow us to target probes to areas where multiple polymorphic residues between homologhomologs exist within a small region or to areas where variant sites occur with high frequency in the population.
We also found that the number of haptens present on a probe can influence the signal intensity and therefore the utility of such probes. The size of alpha satellite DNA arrays vary widely, both between different chromosomes and between homologs. During our studies, we identified a chromosome 15 that had an alpha satellite DNA array so small that it was below the sensitivity level of our oligonucleotide probes. Within intermediate-sized arrays, each sequence variant may be represented by a few hundred or fewer higher-order repeat units. Therefore, the addition of a single hapten to an oligonucleotide probe may mean the difference between a visible signal and no signal at all. Maximizing the number of haptens on an oligonucleotide probe also reduces the number of signal amplification steps necessary for visualization and, subsequently, reduces nonspecific background.
In both this and our previous work (O'Keefe et al. 1996), we identified three general classes of alpha satellite DNA arrays: those with repeats that contain variant base X, those with repeats that contain variant base Y, and those that contain some combination of the two repeats. This three-allele system is a simplification of the true number of alleles present at the centromere. For the pTRA-20 class of chromosome 15-specific alpha satellite DNA, the ratio of repeats containing T at position 529 versus those containing C most likely varies between chromosomes we have scored as being of the T/C class. However, our technique cannot distinguish alleles that have different T/C ratios. An exception is arrays that contain < 100 copies of a particular repeat. Because we are unable to detect sequences that are present in < 100 copies (see below), these arrays would be scored as being negative for that repeat. Therefore, as we have seen for chromosome 15, it is possible that the “heterozygous” class of chromosomes may be overestimated.
Sequence variant-specific oligonucleotide probes are potentially valuable tools for both clinical uses as well as for basic research. Previously, we described oligonucleotide probes specific for chromosome 17 alpha satellite DNA variants. In the present report, we demonstrate that this technique can be extended to chromosome 15 and, ultimately, to all human chromosomes. This technique will not only benefit from sequence data from the other chromosomes but also from the development of more sensitive detection strategies. At present, the sensitivity of established detection protocols has limited this technique to variants that are present in > 100 copies (Matera and Ward 1992; C. O'Keefe, unpubl.). New probe designs and detection strategies, such as padlock probes (Nilsson et al. 1994, 1997), rolling circle amplification (Lizardi et al. 1998), and tyramide deposition (Bobrow et al. 1989; Kerstens et al. 1995; Raap et al. 1995), greatly increase the number of fluorescent molecules that can be deposited at the site of hybridization without sacrificing probe specificity. Techniques such as these may allow future investigators to exploit sequence variants that occur in far fewer than 100 higher-order repeats. Ultimately, we should be able to design a large panel of variant-specific probes for each chromosome, specific for any variant site, increasing the likelihood that a probe pair will be able to distinguish the chromosome homologs.
Variant-Specific Oligonucleotide Probes
Oligonucleotide probes similar to those presented here and in our earlier work (O'Keefe et al. 1996) can be employed to address a number of issues both in basic research and in clinical diagnostic applications. For example, we have used similar probes to study nondisjunction in male meiosis (O'Keefe et al. 1997). Because they are capable of distinguishing homologhomologs, such probes enable us to determine at which meiotic stage a nondisjunction event occurred for informative individuals. Our initial study focused on normal meiosis; further studies examining the effect of environmental exposure to radiation, toxins, or other insults to chromosome- and stage-specific nondisjunction are a logical extension of this study. Such probes could also be used to investigate whether the presence of certain alphoid sequence variants predisposes a chromosome to nondisjunction.
Oligoprobes such as the ones reported here would also be extremely valuable in a clinical genetics facility. Small marker chromosomes can have significant phenotypes, depending on the chromosome from which they were derived, as well as parent of origin. Due to their small size, it is often difficult to determine the chromosome or parent of origin of a marker chromosome. Variant-specific oligonucleotide probes could be used as a first-line technique to identify the chromosome and parent of origin of the centromere. Then, further studies could determine the amount of euchromatic (and theoretically, coding) sequence present in the marker.
Many existing techniques are capable of distinguishing one allele from another. Polymorphic markers in noncentromeric locations are used extensively to create haplotypes for mapping of genes, parent of origin studies, and so forth. However, FISH can be used to address many questions that cannot be answered with test tube-based techniques. One advantage of in situ hybridization techniques is that they can be used on intact cell and tissue preparations. In vitro techniques require that the samples be treated to release the DNA, losing any information about the spatial organization within or between cells. FISH is a nonradioactive technique and can be performed in a single day when using oligonucleotide probes on premade samples. Finally, alpha satellite DNA variant-specific oligoprobes determine the identity of the centromere directly. In contrast, traditional polymorphic markers lie at some distance from the centromere, and therefore, the identity of the centromere must be inferred from the identity of the markers. Occasionally, a chromosome may be assigned the wrong identity if the marker lies far enough from the centromere that a recombination event occurs between them.
The development of oligoprobes capable of distinguishing normal homologous chromosomes by FISH for two chromosomes underscores the fact that this technique can be extended to the remainder of the human chromosome complement. The probes are also capable of distinguishing homologs that vary at a single position, allowing for the development of probes for alpha satellite DNA subfamilies that have limited amounts of sequence variation, such as the subfamily shared by chromosomes 13 and 21 (Devilee et al. 1986; Jorgensen et al. 1987; Vissel and Choo 1992). The development of other such probes is restricted solely by the lack of chromosome-specific alpha satellite DNA sequence information. With the acquisition of such sequence information, the full power of sequence-specific oligonucleotide probes can be achieved.
METHODS
Cloning and Sequencing of Chromosome 15 Alpha Satellite DNA
Plasmid clone pTRA-20 (gift of K.H. Choo) was sequenced from the pUC8 vector primers M13 (-20) Forward and M13 Reverse (Sequinet, Colorado State University). Somatic cell hybrids, monochromosomal for human chromosome 15, were used as a source of DNA for PCR and Southern blot analysis. Cell lines A15, A9 + 15, and GAR-1 contained only human chromosome 15, while cell lines A59–3AZ10a, t60–14, 20L-28, ALA-8, and t75–2maz contained multiple human chromosomes, including a single chromosome 15 (cell lines were gifts of R. Nicholls). DNA was isolated from the cell lines by standard techniques. The sequences of the primers used were as follows: 15SAT7 (5′-GCTTTGAGGATTTCTTTGAAAC), 15SAT8 (5′-TTCAAACCTGCTGTATGAACC), 15SAT9 (5′-CAGGTTTGAATCACTATTTC), 15SAT10 (5′-TACACACATCACAAAGAGC), 15SATb7 (5′-CAAAGTTTCTTTAGGAGTTTCG), 15SATb8 (5′-GGTTCATACAGCAGGTTTGAA), 15SATb9 (5′-CTTTATCACTAAGTTTGGAC), and 15SATb10 (5′-GCTCTTTGTGATGTGTGTA). Approximately 200 ng of DNA was amplified in a 100-μL reaction (200 mM MgCl2, 20 mM dNTP, 100 mM primer; Gibco Taq DNA Polymerase) under the following conditions: 95°C for 2 min; 94°C for 30 s, 59°C (15SAT7/15SAT8) or 53°C (15SAT9/15SAT10, 15SATb7/15SATb10, 15SATb8/15SATb9) for 30 s, 72°C for 30 s (30 cycles); and 72°C for 8 min. The products were gel purified using the Gene Clean kit (Bio101). 15SAT7 was end labeled with 33P ATP using T4 polynucleotide kinase (New England BioLabs) and the purified products were cycle sequenced using the Gibco BRL cycle sequencing kit at 61°C. The sequencing reactions were run on a 4% acrylamide gel. Individual PCR products were cloned from unpurified products into the pCR2.1 vector using the TA Cloning kit (Invitrogen). The clones were cycle sequenced from end-labeled vector primers (M13 [-20] Forward and M13 Reverse primers) as described above.
Preparation of Oligonucleotide Probes
The oligonucleotides used in this study were 15VAR1.1 (5′-GAATCTTCCTTGTGATGTATGCC), 15VAR2.1 (5′-GAATCTTCTTTGTGATGTATGCC), 15VAR1.2 (5′-GAATCTTCCTTGTGAT), 15VAR2.2 (5′-GAATCTTCTTTGTGAT), 15VAR1.3 (5′-TCCT*TGTGATGT*ATGCCCTCAAT*T), 15VAR2.3 (5′-TCTT*TGTGATGT*ATGCCCTCAAT*T, 15VAR1.4 (5′T*CTTCCTTGT*GATGTAT*GCCCT*C-3′), 15VAR2.4 (5′-T*CTTCTTTGT*GATGTAT*GCCCT*C-3′), 15VAR1.5 (5′-T*CAGAAT*CTTCCT*TGT*GAT), and 15VAR2.5 (5′- T*CAGAAT*CTTCTT*TGT*GAT). 15VAR1.1, 15VAR2.1, 15VAR1.2, and 15VAR2.2 were labeled with digoxigenin at the 3′ end using terminal transferase. Primary amine groups were introduced at specific T residues (marked with asterisks) during synthesis. The oligonucleotides were labeled at these sites with either biotin or digoxigenin as described previously (Matera and Ward 1992).
Southern Blot Analysis and Oligonucleotide Probe Stringency Analysis
Standard techniques of DNA analysis were used. pTRA-20 was labeled with 32P using the Rediprime II kit (Amersham). Hybridization was performed in RapidHyb (Amersham) at 65°C for 2 hr. The blots were briefly rinsed in 2XSSC/0.1% SDS at RT, followed by stringency washes (2 × 20 min in 0.1XSSC/0.1% SDS at 65°C).
Plasmid clones p15/78E2 and p15/78F1 were digested with BamH1 to linearize the plasmids and transferred to nitrocellulose membranes by standard procedures (Sambrook et al. 1989). 15VAR1.4 and 15VAR2.4 were end labeled with 33P using T4 polynucleotide kinase (New England BioLabs). Hybridization of 15VAR1.4 and 15VAR2.4 was performed in 30% formamide/2XSSC at 37°C for 2 hr, and stringency washes (3 × 5 min in 30% formamide/2XSSC) were performed at 37°C.
In Situ Hybridization
Metaphase chromosome spreads from normal human peripheral blood lymphocytes, normal lymphoblast cell lines (CEPH families), and mouse/human somatic cell hybrid cell lines were prepared by standard techniques. The slides were pretreated in RNase (100 mg/mL in 2XSSC) at 37°C for 1 hr and then in pepsin (25 μL of 10% stock diluted in 50 mL 0.01M HCl) at 37°C for 10 min, followed by fixation in formaldehyde (1% in 1XPBS/50 mM MgCl2) for 10 min, air drying and dehydrating in an ethanol series (70%, 90%, 100% for 3 min), and air drying again.
Initially, we found that if the oligoprobes were hybridized simultaneously to a single slide, the hybridization pattern differed from when the probes were hybridized individually to independent slides. For example, we found that 15VAR2.4, in the presence of 15VAR1.4, hybridized to chromosomes that were initially scored as negative when 15VAR2.4 was hybridized alone. To overcome this unexpected interaction, we hybridized and detected (see below) the chromosome 15 variant-specific oligoprobes sequentially.
After pretreatment and fixation, the preparations were denatured in 70% formamide/2XSSC at 80°C for 2 min and immediately dehydrated in an ice-cold ethanol series (70%, 90%, and 100% EtOH washes for 3 min) and air dried. Labeled oligonucleotides (∼ 2 ng/μL), along with unlabeled competitor DNA (∼ 600 ng/μL) were hybridized for 1 h at 37°C in 30% formamide/2XSSC/10% dextran sulfate. Two sets of washes (3 × 3 min) were performed as follows: 30% formamide/2XSSC at 37°C followed by 0.1%Tween/2XSSC at 37°C. Detection was performed with conjugated avidin or antidigoxigenin Fab fragments followed by two additional rounds of signal amplification. Image acquisition was performed as described previously (Matera et al. 1995).
Acknowledgments
We thank K.H. Choo for the pTRA-20 clone, R. Nicholls for the chromosome 15 hybrid cell lines, and S. Schwartz for the metaphase cell pellets used in the family transmission studies. This study was funded in part by grant 6-FY98–0637 from the March of Dimes Birth Defects Foundation (to A.G.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL gxm26{at}po.cwru.edu; FAX (216) 368-3432.
-
- Received March 6, 2000.
- Accepted June 15, 2000.
- Cold Spring Harbor Laboratory Press