An EST-enriched Comparative Map of Brassica oleracea and Arabidopsis thaliana
Abstract
A detailed comparative map of Brassica oleracea andArabidopsis thaliana has been established based largely on mapping of Arabidopsis ESTs in two Arabidopsis and four Brassica populations. Based on conservative criteria for inferring synteny, “one to one correspondence” betweenBrassica and Arabidopsis chromosomes accounted for 57% of comparative loci. Based on 186 corresponding loci detected inB. oleracea and A. thaliana, at least 19 chromosome structural rearrangements differentiate B. oleracea andA. thaliana orthologs. Chromosomal duplication in the B. oleracea genome was strongly suggested by parallel arrangements of duplicated loci on different chromosomes, which accounted for 41% of loci mapped in Brassica. Based on 367 loci mapped, at least 22 chromosomal rearrangements differentiate B. oleracea homologs from one another. Triplication of some Brassica chromatin and duplication of some Arabidopsis chromatin were suggested by data that could not be accounted for by the one-to-one and duplication models, respectively. Twenty-seven probes detected three or more loci in Brassica, which represent 25.3% of the 367 loci mapped inBrassica. Thirty-one probes detected two or more loci inArabidopsis, which represent 23.7% of the 262 loci mapped inArabidopsis. Application of an EST-based, cross-species genomic framework to isolation of alleles conferring phenotypes unique to Brassica, as well as the challenges and opportunities in extrapolating genetic information from Arabidopsis toBrassica and to more distantly related crops, are discussed.
Arabidopsis thaliana, a weed-like member of the Cruciferae family (tribe Sisymbrieae), offers many advantages for basic and applied plant research. These features include small stature, short life cycle, small genome size (2n=10, estimated physical genome size of 100–120 Mb), low frequency of repetitive sequences (∼10% of the nuclear genome; Leutwiler et al. 1984), and prolific seed production. These features, combined with research of the past several decades yielding many mutants, efficient transformation systems, detailed genetic and physical maps, the availability of several P1, YAC, and BAC libraries, and 36,569 public ESTs (http://www.cbc.umn.edu/ResearchProjects/Arabidopsis), makeA. thaliana an ideal model for further molecular and genetic study (Meyerowitz and Somerville 1994). A multinational genome research initiative aiming to completely sequence the Arabidopsisgenome by year 2004 (The Multinational Science Steering Committee 1997) is ahead of schedule. Such an accomplishment will undoubtedly create new scientific challenges and opportunities. One of the core issues will be how to apply the information obtained from theArabidopsis genome project to the improvement of the world's leading crops.
The genus Brassica (tribe Brassiceae), including many important crops, is in the same taxonomic family as Arabidopsis thaliana. Such a close relationship suggests that crop plants of the genus Brassica will be among the earliest beneficiaries of a complete sequence of Arabidopsis. Economically,Brassica can be loosely categorized into oilseed, vegetable, and condiment crops. Brassica campestris, Brassica juncea,Brassica napus, and Brassica carinata provide ∼12% of the world-wide edible vegetable oil supplies (Labana and Gupta 1993) and generate >$8 billion market value in North America and Europe. Brassica oleracea and B. campestris, the so-called “Cole crops,” comprise a large variety of vegetables in our daily diet. Many of these vegetables have extreme morphological characteristics of basic interest, such as the enlarged inflorescence of cauliflower (B. oleracea subsp. botrytis) and broccoli (B. oleracea subsp. italica); enlarged stem of kohlrabi (B. oleracea subsp. gongylodes) and marrowstem kale (B. oleracea subsp. medullosa); enlarged root of turnip (B. campestris subsp.rapifera); enlarged and twisted leaves of Pak-choi (B. campestris subsp. chinesis) and Chinese cabbage (B. campestris subsp. pekinesis); and enlarged single apical bud of cabbage (B. oleracea subsp. capitata) or many axillary buds of Brussels sprouts (B. oleracea subsp.gemmifera) (Kalloo and Bergh 1993). Notably, althoughArabidopsis is considered a close relative toBrassica, none of these phenotypes occur inArabidopsis to nearly the same degree. Finally, Brassica nigra is primarily used as a condiment (mustard seed).
Through cytological study, the species relationship of crop Brassicas was described by the “triangle of U” (U 1935). Three allotetraploids, B. juncea (2n=36, AABB), B. napus (2n=38, AACC), and B. carinata(2n=34, BBCC), originated through interspecific hybridization between different pairs of the three diploid species,B. nigra (2n=16, BB), B. oleracea(2n=18, CC), and B. campestris (2n=20, AA). Based on cytological examination and hybrid analysis, the haploid chromosome number of monogenomic species in the Brassiceae were found to range from 7 to 12 (Mizushima 1980). However, because of the available resolution of cytological techniques, detailed genomic relationships among monogenomic species were not fully revealed. Understanding the genomic relationship among monogenomicBrassica species will not only shed light on the evolution of the Brassica genome but also facilitate gene transfer amongBrassica species. The rise of comparative mapping, the alignment of chromosomes based on common DNA markers, has provided the means to study in depth the parallels in genome structure and function of closely related species (Tanksley et al. 1988; Ahn and Tanksley 1993), and distantly related species (Paterson et al. 1996).
The present study aimed to better characterize the comparative genome organization of Brassica and Arabidopsis. Previous study of the genus Brassica showed that the proportion of low-copy DNA sequences was similar among diploid Brassicaspecies, but a large number of rearrangements result in distinct chromosomal number and organization (Slocum et al. 1990; Landry et al. 1991, 1992; Song et al. 1991; Kianian and Quiros 1992; Lagercrantz and Lydiate 1996). Corresponding chromosomes in diploid and amphidiploidBrassica have been reported (Teutonico and Osborn 1994; Cheung et al. 1997a,b). Comparative mapping between Arabidopsis andBrassica revealed even more extensive chromosomal rearrangements (Kowalski et al. 1994a; Lagercrantz et al. 1996; Osborn et al. 1997). These studies, however, did not provide a complete scope of the genome comparison between Brassica andArabidopsis because of the limited numbers of common markers. To address this issue, a larger number of markers were needed on the comparative maps. The present work, based on 186 corresponding loci detected, provides a much more detailed picture of the comparative genome organization of B. oleracea and A. thaliana. Furthermore, study of chromosomal duplication within the B. oleracea genome, based on 367 loci, illustrates some of the complexities that will be faced both in extrapolating Arabidopsisinformation to Brassica and in assembly of sequence-ready contigs for crop genomes.
RESULTS
DNA Polymorphism
Table 1 summarizes the DNA polymorphism detected by 200 Arabidopsis EST clones and 123 Brassica PstI genomic clones. The relatively low level of polymorphism in the B. oleracea (RCB)×B. oleracea ssp. alboglabra var.Bugh Kana (BK) F2 population was consistent with the origin of RCB fromB. oleracea ssp. alboglabra types (Song and Osborn 1992). The chance of detecting polymorphic probes is low and similar in bothArabidopsis crosses. There is variation in polymorphism rate associated with different restriction enzymes in Brassica, but no particular pattern is clear. In Arabidopsis, the restriction enzyme CfoI consistently detects more polymorphism than the other restriction enzymes, in both populations.
Summary of the Polymorphism Detected by Arabidopsis EST Clones and Brassica PstI Genomic Clones
Establishing Composite Linkage Maps
B. oleracea Linkage Maps
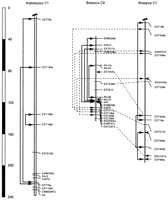
Because many of the mapped polymorphisms were unique to one B. oleracea population, we constructed a composite linkage map forB. oleracea to more completely reflect all of the available comparative information. The assembly of the B. oleraceachromosome 1 composite map was illustrated in Figure1 as an example, built according to the following rules: (1) Common loci detected in different populations could be identified based on the size of the restriction fragment from RCB, the common parent. These permitted the initial alignment of chromosomes of different populations. (2) The RCB×GC map was used as the primary linkage map because the largest number of loci were mapped in this population. Markers that did not detect polymorphism in RCB×GC population but did detect polymorphism in other populations were mapped in other populations accordingly. For chromosome 8, where the RCB×GC map exhibited few polymorphic markers, the RCB×PK linkage map was substituted. (3) The integration of unique loci was based on the closest common flanking loci, and the unique loci were positioned proportionally to their proximity to the flanking loci. (4) To test possible chromosomal rearrangements in different varieties, lod scores were calculated for the alternative (consensus) orders. Only if each possible consensus order in both populations could be ruled out by lod 2.0 was a rearrangement suggested.
The assembly of the Brassica chromosome 1 composite map. Common loci (based on common restriction fragment sizes) were connected by solid lines, putatively homologous loci (with different restriction fragment sizes, but at corresponding sites) are connected by dashed lines. Filled circles placed on crossed lines indicate that respective orders of loci are statistically significantly different (≥LOD 2.0) in the respective maps, suggesting possible chromosomal rearrangements. Arrows indicate the inferred locations of unique loci in the consensus map.
The linkage maps span recombinational lengths of 743.0, 893.2, 947.1, and 871.3 cM across the B. oleracea genome in RCB×GC, PK, CAN, and BK populations, respectively, with an average length of 863.6 cM. The average recombinational lengths of B. oleraceachromosomes 1–9 are 189.2, 102.4, 91.7, 95.7, 97.4, 83.4, 77.2, 71.9, and 72.9 cM, respectively. A total of 367 loci were detected in the composite map with an average interval between loci of 2.35 cM. Based on an estimated DNA content of 660 Mb (Arumuganathan and Earle 1991), this corresponds to an average spacing of 1.8 Mb between markers and suggests that most genes are within 0.9 Mb of the nearest marker. Table 2 summarizes possible chromosomal rearrangements found among different B. oleracea varieties.
Possible Chromosomal Rearrangements Detected in DifferentBrassica Varieties
A. thaliana Linkage Maps
Construction of the A. thaliana composite map (Fig. 3, below) has been reported previously (Kowalski et al. 1994a), although this report includes 152 more loci. Specifically, common DNA polymorphisms detected on both populations served as “anchor loci” to infer the relative order of loci segregating in only one of the two populations. The map includes 262 loci across the A. thalianagenome. Thirty-one probes detect duplicated loci. Table3 summarizes 20 duplicated loci newly detected by ESTs and cloned RAPD-amplified genomic DNA.
Composite RFLP linkage map of Arabidopsis thaliana HM x WS and M13 x WS F2 populations. Markers designated “FQ” are anchor loci, common to both populations. Markers mapped in the HM x WS population are designated “Q”. The remaining markers were mapped in M13 x WS only. The construction of the A. thaliana composite map was as reported previously (Kowalski et al. 1994a). It should be noted that integration of data from two populations tends to inflate recombinational distance due to unequal recombination between populations. The filled circles next to the loci indicate homoeologous loci detected by the same probe on the Brassica composite map. Open circles indicate that no polymorphism was detected for homoeologous loci in RCB x GC Brassica populations. The letter “R” next to a probe name indicates that it hybridizes to a repetitive DNA sequence in the Brassica genome. Specific colors are asigned to each homoeologous chromosome. Markers included in the one-to-one model for Arabidopsis-Brassica correspondence are connected by filled columns. Open columns indicate possible duplicated regions in Brassica.
Summary of Newly Detected Duplicated Loci in Arabidopsis Genome
Patterns of Correspondence of Brassica Chromosomes with One Another and with the Arabidopsis Chromosomes
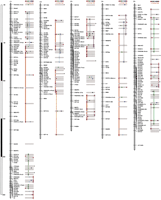
Figure 2 illustrates and Table4 summarizes the composite linkage map of B. oleracea. We developed a model for the comparative organization of the chromosomes of B. oleracea and A. thaliana that assumes duplication of most Brassica chromosomes and one-to-one correspondence of Brassica chromosomes withArabidopsis chromosomes. The extent to which the observed data cannot be explained by this “null hypothesis,” reflects the need for alternative hypotheses such as triplication of Brassicachromosomal segments or duplication of Arabidopsis chromosomal segments. The model was built based on the identification of SCEUS (smallest conserved evolutionaryunit segments; O'Brien et al. 1993) of three or more loci that (1) maximize the number of corresponding DNA marker loci that are consistent with the model, (2) minimize the number of chromosomal rearrangements between duplicates (Brassica) or orthologs (Arabidopsis), (3) consider closely linked markers to be stronger evidence of synteny than distantly linked markers, and (4) consider a genetic distance of >5 cM to represent a true difference in locus order. This relatively large value was chosen to reflect not only the small size of the primary population but also the uncertainties associated with inference of loci mapped in other populations. Further constraints were imposed to evaluate the extent of duplication and triplication in Brassica. Specifically, possible regions of duplication along a chromosome were inferred first, in a manner that followed the above rules and did not allow different duplicated segments to overlap with each other by >5 cM (the threshold for inferring rearrangement). Finally, regions of possible triplication were inferred: These were allowed to overlap with duplicated segments but not with each other. From first principles, if the duplication process in Brassica were random (not associated with large chromosomal region), the duplication model would explain 12.5% of data (given that B. oleracea has nine chromosomes). The extent to which the model improves on this reflects the strength of evidence for duplication and triplication. By the same rationale, one-to-one correspondence of Brassica toArabidopsis must account for significantly more than the random expectation of 25% of data to be meaningful. Higher levels of correspondence in small chromosomal regions may be suggestive of duplication of chromosomal segments.
Composite RFLP linkage map of Brassica olearacea RCB x GC, RCB x CAN, RCB x PK and RCB x BK F2 Populations. Filled circles next to the loci indicate homoeologous Brassica loci (chromosomes 1-9, near right) or homologous Arabidopsis loci (chromosomes 1-5, far right) detected by the same probe. Open circles indicate that no polymorphism was detected for homoeologous (Brassica) and homologous (Arabidopsis) loci. A letter “R” next to the probe name indicates that the probe hybridizes to a repetitive DNA sequence in Arabidopsis. Specific colors are assigned to each homoeologous and homologous chromosome. Markers included in the duplication (Brassica) or one-to-one (Arabidopsis) models are connected by colored columns. Open columns indicate possible triplicated (Brassica) or duplicated (Arabidopsis) regions.
Summary of Brassica–Brassica andBrassica–Arabidopsis Correspondence
Brassica Chromosome 1
The “duplication” model, in which Brassica chromosome 1 corresponds to nonoverlapping segments of Brassica chromosomes 4, 9, and 6 (sequentially, moving down the chromosome), explains only 40% of the additional loci detected by probes for which at least one locus mapped to chromosome 1. Loci that are not included in the duplication model occur in several closely linked clusters that suggest higher order redundancy of chromatin. In particular, 20 loci suggest correspondence to regions of chromosomes 7 (near top), 5 and 3 (nonoverlapping regions near middle), 4 (parallel to upper part of chromosome 6 correspondence), and 8 and 1 (nonoverlapping regions parallel to lower part of chromosome 6 correspondence), which represent possible “triplicated” chromosomal segments and account for 23% of the corresponding loci. Eight additional loci corresponding to chromosomes 3 and 9 (near the bottom) are noted but could not be inferred to be syntenic by the rules of our model.
One-to-one correspondence to regions of Arabidopsischromosomes 5, 4, 3, and 1 (moving down the Brassicachromosomes) accounts for 47% of corresponding loci. Possible duplication in Arabidopsis is suggested by five loci corresponding to Arabidopsis chromosomes 2 (parallel to chromosome 5 correspondence) and 8 (parallel to chromosome 4 and 1 correspondence), accounting for 26% of the corresponding loci.
Brassica Chromosome 2
One-to-one correspondence suggests an internal duplication where the upper part of the chromosome (EW7B04b–EW7B04c) corresponds to the lower part of the chromosome (EW6A04b–EW4D12c), based on nine loci. The middle of chromosome 2 corresponds to chromosomes 8 and 4. Overall, these data explain 55% of the duplicated loci. Loci that are not included in this model suggest two possible segments corresponding to chromosomes 6 and 1 (parallel to chromosome 4 correspondence) and explain 21% of the corresponding loci.
One-to-one correspondence to regions of Arabidopsischromosomes 2 and 5 accounts for 54% of corresponding loci. Three additional loci on Arabidopsis chromosome 2 partially overlap the correspondence of Arabidopsis chromosome 5.
Brassica Chromosome 3
One-to-one correspondence to segments of Brassicachromosomes 5, 1, and 8 explains 38% of the duplicated loci. Loci that are not included in this model suggest a triplicated segment corresponding to chromosome 6 (parallel to chromosome 1 correspondence) and explain 14% of the corresponding loci. Three isolated loci corresponding to chromosome 4 are noted but cannot be accommodated by the rules of the model.
One-to-one correspondence to regions of Arabidopsischromosomes 1 and 3 accounts for 69% of the corresponding loci.
Brassica Chromosome 4
One-to-one correspondence to a segment of Brassicachromosome 1 explains 43% of the duplicated loci. Loci that are not included in this model suggest triplicated regions corresponding to chromosome 5, 3, 6, and 7, which explain 17% of corresponding loci. Additional loci corresponding to chromosome 6 and 8 are noted but cannot be accommodated by the rules of the model.
One-to-one correspondence of Brassica chromosome 4 toArabidopsis chromosome 5 explains 43% of the corresponding loci. Loci that are not included in this model suggest duplicated regions correspond to Arabidopsis chromosome 1 and explain 27% of corresponding loci. Additional loci corresponding to chromosome 3 are noted but cannot be accommodated by the rules of the model.
Brassica Chromosome 5
One-to-one correspondence to Brassica chromosome 1 explains 32% of the duplicated loci. Three loci not included in this model suggest a triplicated region corresponding to chromosome 9 explaining 12% of the corresponding loci. Four loci corresponding to chromosome 4 are noted but cannot be accommodated by the rules of the model.
One-to-one correspondence to regions of Arabidopsischromosomes 1 and 2 explains 73% of the data.
Brassica Chromosome 6
One-to-one correspondence to segments of Brassicachromosomes 1 and 4 explains 44% of the duplicated loci. Loci that are not included in this model suggest a triplicated region corresponding to chromosome 2 and explain 9% of the corresponding loci. Isolated loci corresponding to chromosomes 2 and 8 are noted but cannot be accommodated by the rules of the model.
One-to-one correspondence to regions of Arabidopsischromosomes 1 and 2 explains 57% of the corresponding loci. Three loci corresponding to chromosome 4 are noted.
Brassica Chromosome 7
One-to-one correspondence to segments of Brassicachromosomes 1 and 9 explains 42% of the duplicated loci. Loci that are not included in the model suggest a triplicated region corresponding to chromosome 4 and explain 16% of corresponding loci.
One-to-one correspondence to a region of Arabidopsischromosome 5 explains 33% of the corresponding loci.
Brassica Chromosome 8
One-to-one correspondence to segments of Brassicachromosomes 4 and 3 explains 33% of the duplicated loci. Loci that are not included in this model suggest a triplicated region corresponding to chromosome 1 and explain 21% of corresponding loci. Four loci corresponding to chromosome 2 and three loci corresponding to chromosome 6 are noted.
One-to-one correspondence to regions of Arabidopsischromosomes 4 and 3 explains 80% of the corresponding loci.
Brassica Chromosome 9
One-to-one correspondence suggests an internal duplication of chromosome 9 where the chromosomal segment EW8E09d– AKJ2c corresponds to the segment AKJ2b–K457b, involving 10 loci. An intervening region corresponds to chromosome 7, and the lower part of chromosome 9 corresponds to chromosome 1. Overall, these regions explain 46% of the duplicated loci. Loci that are not included in this model suggest triplicated regions corresponding to chromosome 5 and explain 16% of corresponding loci. Four loci corresponding to chromosome 8 and three loci corresponding to chromosome 6 are noted but cannot be accommodated by the rules of the model.
One-to-one correspondence to regions of Arabidopsischromosomes 1 and 5 accounts for 81% of the corresponding loci. Three loci correspond to Arabidopsis chromosome 3 are parallel to chromosome 5 correspondence and may reflect duplication in Arabidopsis.
Patterns of Correspondence of Arabidopsis Chromosomes with the Brassica Chromosomes
The “one-to-one correspondence” model and duplication model were also tested on the Arabidopsis linkage map as well, which was illustrated in Figure 3 and summarized in Table5.
Summary of Arabidopsis–Brassica Correspondence
Arabidopsis Chromosome 1
The one-to-one model, in which Arabidopsis chromosome 1 corresponds to nonoverlapping segments of Brassica chromosomes 5, 1, 4, and 9, explains 46% of the loci detected by probes for which at least one locus mapped to chromosome 1. Loci that are not included in the one-to-one model suggest a duplicated region corresponding toBrassica chromosome 3 (parallel to chromosome 5 correspondence), explaining 10% of corresponding loci. Three loci corresponding to Brassica chromosome 6 are noted.
Arabidopsis Chromosome 2
The one-to-one model, in which Arabidopsis chromosome 2 corresponds to segments of Brassica chromosomes 1, 5, 1, and 2, explains 52% of the loci. Loci that are not included in the model suggest a duplicated region corresponding to chromosome 6, explaining 15% of corresponding loci.
Arabidopsis Chromosome 3
Our model suggests the correspondence of Arabidopsischromosome 3 to nonoverlapping segments of Brassicachromosomes 1, 8, and 1 sequentially, explaining 49% of the duplicated loci. Loci that are not included in the model suggest duplicated segments of chromosome 4 (near top) and 9 (near bottom), explaining 17% of corresponding loci.
Arabidopsis Chromosome 4
The model suggests the correspondence of Arabidopsischromosome 4 to Brassica chromosomes 8 and 4 and explains 36% of the loci. Loci that are not included in the model suggest a duplicated region corresponding to chromosome 6, explaining 14% of corresponding loci. Three loci corresponding to chromosome 7 are noted.
Arabidopsis Chromosome 5
Our model suggests the correspondence of Arabidopsischromosome 5 to segments of Brassica chromosomes 4, 9, and 4 and explains 39% of the corresponding loci. Loci that are not included in the model suggest duplicated regions corresponding to chromosome 1, explaining 23% of the corresponding loci. Six loci corresponding to chromosome 7 are noted.
DISCUSSION
It is timely to consider the challenges and opportunities in extrapolating structural genomic information from Arabidopsis, the first plant for which the genome will be completely sequenced, toBrassica and other more distantly related plants.
Our model (Fig. 2) suggests that at least 22 chromosomal rearrangements differentiate the B. oleracea homologs from one another and at least 19 rearrangements differentiate A. thaliana from B. oleracea. In several instances the locations of chromosomal rearrangement breakpoints between Brassica homologs approximately match the locations of the breakpoints betweenArabidopsis and Brassica. Some such instances include (1) Brassica chromosome 2, where the correspondence withBrassica chromosomes 2 and 8 breaks between EW7B04c and EW6G12a and the correspondence with Arabidopsis chromosomes 2 and 5 breaks between EW1F08 and EW2E05b; (2) Brassicachromosome 3, where the correspondence with Brassicachromosomes 1 and 8 breaks between EST130a and EW2D03a and the correspondence of Arabidopsis chromosomes 1 and 3 breaks between EW7D03y and EW2D03a; (3) Brassica chromosome 8, where the correspondence with Brassica chromosome 4 and 3 homologs breaks between EW5G04b and EST517d and the correspondence ofArabidopsis chromosomes 4 and 3 breaks between EST22a and EW8F03b; (4) Brassica chromosome 9, where the correspondence of Brassica chromosomes 9 and 1 breaks between K457b and EST517g and the correspondence of Arabidopsis chromosomes 1 and 5 breaks between EW1G03a and EST9a. Such rearrangement breakpoints that appear to be common to Brassica and Arabidopsismay reflect cases where both Arabidopsis and oneBrassica homolog retain the chromosome organization of their common ancestor, whereas a duplicated Brassica homolog has undergone rearrangement. Similarly, chromosomal regions in whichArabidopsis gene order corresponds to one but not bothBrassica homoeologs may reflect rearrangement of oneBrassica homoeolog since duplication. For example, onArabidopsis chromosome 5, the order for marker EW5D12, EST075, and EST150 is EST150–EST75–EW5D12, and on Brassicachromosome 4, it follows the same order, but on Brassicachromosome 1, the order changes to EST75–EST150–EW5D12.
Comparative Organization of Brassica Homoeologous Chromosomes
The Brassica chromosomal duplication model explains 41% of the duplicated restriction fragment length polymorphism (RFLP) loci we mapped (Table 4). If there were no pattern to duplication, then the duplication would be expected to account for <12.5% (1 out of 8) of data, because there are nine pairs of chromosomes in B. oleracea.Our data clearly indicate that duplication has involved large chromosome segments in Brassica. In a similar manner, if triplication accounts for more than an additional 14.3% (1 out of 7) of data in Brassica, then it would be more common than expected to occur at random. Based on our model, triplication ofBrassica chromosomal segments best explains 18% of the data, which is nominally greater than the expected value (14.3%). Although the case for triplication is much weaker than for duplication, the clustering of triplicated loci into linked groups does tend to support prior suggestions based on smaller numbers of probes and isolated genomic regions (Kowalski et al. 1994a; Lagercrantz et al. 1996; Osborn et al. 1997) that some regions of the genome of B. oleracea(as well as B. rapa and B. nigra) may be triplicated. A fundamental problem in the use of genetic mapping data to evaluate duplication (and triplication) of chromatin is the need to detect DNA polymorphism. The assembly of physical maps for the Brassicagenomes will alleviate this limitation but will require new methodology to efficiently determine the locus-specificity of BACs (or other large DNA clones) that hybridize to duplicated (or triplicated) probes.
Alignment of Brassica and Arabidopsis Chromosomes
The Brassica/Arabidopsis one-to-one correspondence model explain 57% of our observed data (Table 4). If the genomes ofBrassica and Arabidopsis were randomly arranged with respect to one another, then one-to-one correspondence would account for < 20% (1 out of 5) of data in Arabidopsis. Our data clearly indicate extensive synteny of Brassica andArabidopsis.
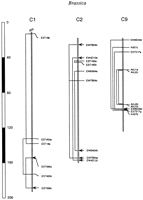
A total of 31 pairs of duplicated loci, including 20 pairs reported here for the first time (Table 3), mapped to A. thaliana, accounting for 23.7% of the loci detected. These duplicated loci expand on the earlier suggestion (Kowalski et al. 1994a) that part of the A. thaliana genome may have undergone ancient duplication. These ancient duplications could complicate contig-map construction and also could reduce the subset of Arabidopsis genes that are susceptible to “knockout” experiments (Sundaresan et al. 1995;Kempin et al. 1997). Notably, an intrachromosomal duplication appears to occur in A. thaliana chromosome 1 (Fig. 4).
Intrachromosomal duplication of Arabidopsis chromosome 1, and an possible more-than-triplicated region of Brassicachromosome 1 and 9. Solid lines connect homoeologous loci (based on different restriction fragment sizes) located on the same chromosome. Dashed lines connect homoeologous loci located on different chromosomes.
Intrachromosomal duplication was observed in chromosomes 1, 2, and 9 ofB. oleracea (Fig. 5). Two independent studies on the genome of B. nigra reveal similar patterns on chromosome 5 (Truco and Quiros 1994) and chromosome 6 (Lagercrantz and Lydiate 1996), suggesting that such intrachromosomal duplication might be common in Brassica. If such intrachromosomal duplications preceded the duplication/triplication of the ancestral B. oleraceagenome, then even higher levels of duplication might be expected in modern B. oleracea. In our study, five probes did detect more than three segregating loci in B. oleracea, including EW4D04 (chromosomes 1, 2, 4, and 8), EW8A06 (chromosomes 1, 4, 5, and 7), EST55 (chromosomes 1, 2, 4, and 6), EST453 (chromosomes 1, 4, 5, 6, and 9), and EST517 (chromosomes 1, 6, 8, and 9). Although we cannot rule out the possibility that some of these more-than-triplicated loci might be the consequence of other duplication mechanisms, segments ofBrassica chromosomes 1 and 9 did suggest the existence of such high-order chromosome segmental duplication (Fig. 5). More probes mapped in this region should provide further evidence.
Through comparative mapping, many powerful tools already created forArabidopsis can now be applied to Brassica. For example, Arabidopsis cDNA sequences may be used to isolate homologous genes in Brassica, Arabidopsis BAC/YAC contigs may be used in Brassica for map-based cloning, andArabidopsis high-resolution maps may help to resolve clustered markers in Brassica (Liu et al. 1996). Arabidopsisgenomic tools may guide the isolation of Brassica alleles conferring unique phenotypes. Brassica andArabidopsis may have diverged as little as 10 mya (Muller 1981), suggesting that ∼90% of chromosomal segments <5 cM may remain colinear (Paterson et al. 1996). A comparative map with a density of <5 cM/marker makes it relatively easy to evaluate correspondence of Brassica quantitative trait loci (QTLs) toArabidopsis mutations or candidate genes. Furthermore, a comparative map of B. oleracea (CC genome) and A. thaliana can be extended to an amphidiploid species ofBrassica such as B. napus (AACC genome), where genome complexity is redoubled.
Genetic linkage maps based on ESTs (Berry et al. 1995) enable one to use sequence information to screen for conservation with distantly related taxa. For example, disease-resistance-like ESTs could be potentially useful in locating disease-resistance loci in a specifically designed segregating population other thanArabidopsis (Botella et al. 1997). Also, through selection of highly conserved ESTs, comparative organization of the chromosomes of even distantly related species such as Arabidopsis,Gossypium (cotton), and Sorghum can be studied using the same probes (Paterson et al. 1996). Thus, a cross-genome comparative map based on a common set of ESTs may eventually provide a direct comparison of macro- and microcolinearity across various species. The combination of ESTs and DNA microarray technology (Winzeler et al. 1998) could accelerate this process. Furthermore, mapping the common set of ESTs to Arabidopsis megabase DNA libraries (Schmidt et al. 1995; Zachgo et al. 1996; Agyare et al. 1997) will extend the Arabidopsis physical map and DNA contigs to other plants. Thus, using Arabidopsis contigs to assist map-based cloning in cotton, sorghum, or other genomes may be more feasible. Such a cross-genome framework and toolbox could profoundly affect future genome sequencing projects in related taxa. It is of interest not only to elucidate the portions of genome that are conserved (common) among various species but also the portions that are divergent among species. Thus, the priority of subsequent crop genome sequencing projects might be focused on genomic regions that are poorly conserved, so that scarce financial resources are used more efficiently.
METHODS
Plant Materials
Two A. thaliana F2 populations were used in this study: A. thaliana ecotype Wassilewskija (WS)×mutant stock M13 (Liu et al. 1996) and WS×Hannover/Münden (HM) (Kowalski et al. 1994b). Subsets of 78 individuals from each population were used for mapping Arabidopsis ESTs. Four B. oleraceaF2 mapping populations were used in this experiment: RCB (self-compatible)×B. oleracea var. Green Comet (USDA collection, accession no. G30771, from North America), RCB×B. oleracea var. Cantanese (USDA collection, accession no. PI462224, originally from Italy), RCB×B. oleracea var.Pusa Katki (USDA collection, accession no. PI274783, originally from India), and RCB×B. oleracea var.Bugh Kana (USDA collection, accession no. PI249556, originally from Thailand), composed of 56, 247, 250, and 246 individuals, respectively. A. thaliana seed were obtained from the Arabidopsis Biological Resources Center at Ohio State University, directed by Dr. R.L. Scholl. Rapid-cyclingBrassica was from the Crucifer Genetics Cooperative, Madison, WI. Seed and pollen of other B. oleracea varieties were generously provided by Dr. J. McFerson and Dr. S. Kresovich, then at USDA–ARS, Geneva, NY.
Genotyping
DNA extraction, electrophoresis, Southern blotting and autoradiography were as described previously (Kowalski et al. 1994a). A total of 113 Brassica PstI genomic clones (“EW,” “WG,” and “WR,” from Pioneer HiBred), 35 Arabidopsisgenomic clones (“M,” from Dr. E. Meyerowitz, Caltech), 23Arabidopsis anonymous cDNA clones (“AC,” “ATEX,” and “TCH”), four cloned RAPD-PCR products (“R;” unpubl.), 198Arabidopsis EST clones (“EST,” from Dr. R.L. Scholl, theArabidopsis Biological Resources Center, Ohio State University), and 19 putatively embryo-specific Arabidopsis EST clones (“AHD,” “AKJ,” “AKN,” “Cla,” “d2P,” “FLS,” “HD,” “HMG,” “K,” “S,” and “Seed,” from Dr. Terry L. Thomas, Texas A&M University) were used in this study.
Data Analysis
RFLP linkage maps were constructed using MapMaker (Lander et al. 1987). Linkage groups were built at threshold of lod (logarithm of odds)=2.1 for A. thaliana and lod=2.5 for B. oleracea. Genetic distances (in centiMorgans) were calculated using the Kosambi mapping function.
Acknowledgments
We thank Tzung-Fu Hsieh for critical discussion, Kenneth Feldmann and JoVan Currie for technical help, the Texas Higher Education Coordinating Board, USDA Plant Genome Program, and Texas Agricultural Experimental Station for funding. We thank Pioneer HiBred Production, Ltd. for providing a subset of the DNA probes used.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
Present addresses: 2USDA–ARS, Beltsville, Maryland 20704 USA; 4Washington Fruit Tree Research Commission, Wenatchee, Washington 98801 USA; 5Department of Plant Breeding and Biometry, Cornell University, Ithaca, New York 14850 USA; 6Applied Genetic Technology Center, Department of Crop and Soil Sciences, Department of Botany, and Department of Genetics, University of Georgia, Athens, Georgia 30602 USA.
-
↵7 Corresponding author.
-
E-MAIL paterson{at}uga.edu; FAX (706) 583-0160.
-
- Received August 18, 1999.
- Accepted March 27, 2000.
- Cold Spring Harbor Laboratory Press