Physical and Genetic Mapping of the Human X Chromosome Centromere: Repression of Recombination
Abstract
Classical genetic studies in Drosophila and yeast have shown that chromosome centromeres have a cis-acting ability to repress meiotic exchange in adjacent DNA. To determine whether a similar phenomenon exists at human centromeres, we measured the rate of meiotic recombination across the centromere of the human X chromosome. We have constructed a long-range physical map of centromeric α-satellite DNA (DXZ1) by pulsed-field gel analysis, as well as detailed meiotic maps of the pericentromeric region of the X chromosome in the CEPH family panel. By comparing these two maps, we determined that, in the proximal region of the X chromosome, a genetic distance of 0.57 cM exists between markers that span the centromere and are separated by at least the average 3600 kb physical distance mapped across the DXZ1 array. Therefore, the rate of meiotic exchange across the X chromosome centromere is <1 cM/6300 kb (and perhaps as low as 1 cM/17,000 kb on the basis of other physical mapping data), at least eightfold lower than the average rate of female recombination on the X chromosome and one of the lowest rates of exchange yet observed in the human genome.
Meiotic exchange is not distributed randomly along the length of eukaryotic chromosomes; indeed, much regional variation in recombination frequency has been observed. Perhaps the most conspicuous departure from uniformity is the dramatic repression of exchange found near eukaryotic chromosome centromeres and some telomeres (Mather 1936, 1939). Repression of meiotic recombination adjacent to the centromere (the centromere effect) is most obvious on the Drosophila X chromosome in which the centric heterochromatin, comprising half of the chromosome’s cytogenetic length, barely contributes to its genetic length (Mather 1939; Roberts 1965); a similar centromere-associated repression of recombination is found on the autosomes of Drosophila (Beadle 1932; Painter 1935; Thompson 1963). Some of this repression of exchange is caused by the large blocks of heterochromatin present at the centromeres of higher eukaryotes (Willard 1990; Murphy and Karpen 1995), because heterochromatin, at least in Drosophila, is a poor substrate for recombination regardless of chromosomal location (Baker 1958). Because deletions of centric heterochromatin result in lowered levels of meiotic exchange in centromere-adjacent euchromatin (Yamamoto and Miklos 1978), the presence alone of heterochromatin atDrosophila centromeres does not fully explain the centromere effect. Rather, the centromere seems to exert a suppression of recombination that spreads to adjacent DNA.
Studies in yeast support a similar centromeric suppression of meiotic exchange in proximal chromosome regions, although this effect may be less pronounced. In both Saccharomyces cerevisiae andSchizosaccharomyces pombe, mitotic recombination is relatively more frequent than meiotic recombination in the proximity of centromeres (Malone et al. 1980; Minet et al. 1980). Direct evidence for the centromere effect in yeast has come from studies of cloned centromeres, in which DNA adjacent to the 60-kb centromere domain ofS. pombe has an apparently lowered rate of meiotic exchange (Nakaseko et al. 1986). Further, in S. cerevisiae, removal and distal displacement of a 600-bp fragment containing the centromere of chromosome III (CEN3) results in a three- to fivefold decrease in crossing over at the new site of integration (Lambie and Roeder 1986). Although the genetic maps of most other higher eukaryotes are less well characterized, there is evidence in Neurospora, corn, tomato, and barley for a centromeric suppression of meiotic recombination (Charlesworth et al. 1986; van Daelen et al. 1993; Davis et al. 1994).
In humans, chiasma distributions predict that crossing over occurs more rarely near chromosome centromeres than elsewhere (Hulten 1974). Until recently, the lack of detailed long-range physical maps spanning centromeres (and uncertainty regarding the functional identification of a centromere itself) has precluded the definitive measurement of recombination rates at human centromeres. Preliminary comparisons of physical with genetic distances at or near the centromeres of several human chromosomes have resulted in various estimates of pericentromeric recombination rates, some consistent (Carson and Simpson 1991; Lichter et al. 1993; Weeks et al. 1995) and others inconsistent (Jabs et al. 1991; Janson et al. 1991; Van Hul et al. 1993) with a suppression of centromeric meiotic exchange. A recent study, which compares a yeast artificial chromosome (YAC) physical map with the corresponding genetic map, suggests that recombination may be reduced by an order of magnitude across the centromere of chromosome 10 (Jackson et al. 1996). Data from recent radiation hybrid (RH) maps are also consistent with a repression of exchange at human centromeres (James et al. 1994; Hudson et al. 1995). These data are difficult to interpret, however, because proximal chromosome regions may be preferentially retained in RH cell lines, at a frequency disproportionate to their physical size (D. Slonim and M.M. Mahtani, unpubl.).
Here, we use the tools of pulsed-field gel mapping to provide an estimate of recombination across the human X chromosome centromere relative to a measured physical distance, using markers that are exclusively chromosome-specific. By comparing long-range physical maps with their corresponding genetic maps, we find that meiotic exchange is repressed at least 8-fold, and perhaps as much as 20-fold, at the centromere relative to the average exchange rate on the human X chromosome.
RESULTS
Physical Map Across the Centromere: Rationale
To determine the physical distance across the centromere, we constructed long-range restriction maps spanning the α-satellite array (DXZ1) at two very disparately sized X chromosome centromeres. α satellite is a repetitive DNA family found at the centromeres of all human chromosomes and has been implicated in centromere function (Tyler-Smith and Willard 1993; Larin et al. 1994; Harrington et al. 1997). It is based on a ∼171-bp monomer unit that is tandemly arranged into long arrays comprising up to several megabases of DNA (Willard 1990). Different chromosomes and different copies of the same chromosome carry often highly variable quantities of α satellite at their centromeres (Wevrick and Willard 1989; Mahtani and Willard 1990;Oakey and Tyler-Smith 1990). On the X chromosome, extending results of an earlier study (Mahtani and Willard 1990), we find that the amount of α-satellite DNA ranges from 2200 to 3730 kb at individual centromeres, with a mean of 3010 kb (s.d. = 429;n = 49). One outlier (3.5 s.d. away from the mean), however, was identified (Mahtani and Willard 1990) with an unusually small DXZ1 array size (∼1500 kb, cell line LT690; see below). Because several X chromosomes from CEPH individuals were included in the size estimates, genetic distances derived in CEPH meioses can be expected to reflect recombination across array sizes of, on average, ∼3010-kb arrays of α-satellite DNA.
Two cell lines, LT690 and AHA–11aB1, a mouse/human somatic cell hybrid segregating a human X chromosome as its only human material, were chosen for long-range mapping on the basis of these array size measurements. AHA–11aB1 contains a single X chromosome with an α satellite array length of ∼3000 kb, very close to the observed mean array length for the X chromosome. In contrast, the male lymphoblast line LT690 has the smallest array size yet measured (Mahtani and Willard 1990). By making two maps, we hoped to compare the structural organization of the full-sized AHA–11aB1 array with the shorter LT690 array.
Structural Similarities Between the Two Arrays: Mapping with Common-Cutting Enzymes
The general features of the restriction site maps of the two X chromosome arrays were similar. Digests with common-cutting enzymes were used to give total size estimates of the arrays, because by definition, these enzymes digest away most nonrepetitive flanking DNA from α-satellite-containing restriction fragments (Wevrick and Willard 1989; Mahtani and Willard 1990; Oakey and Tyler-Smith 1990). These enzyme digests also allowed us to position restriction sites within the array. The derived long-range restriction maps of the two arrays are presented in Figure 1, and representative data for the LT690 array are shown in Figure 2.
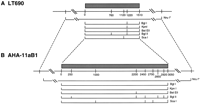
Restriction maps of centromeric α-satellite arrays and adjacent DNA in LT690 (A) and AHA–llaB1 (B). Distances are shown in kilobases; α-satellite sequences are present on those fragments indicated above by hatching. Rare cutting restriction enzymes such asNruI (*) have sites outside the array edges; these enzymes yield restriction fragments containing the entire α-satellite array along with variable amounts of flanking sequence. Fragment sizes obtained by digestion with these restriction enzymes are listed in Table 1, although the positions of these sites relative to one another was not determined. The broken lines point out the similar features of the DNA near the two chromosome centromeres: the clustering of common cutting restriction fragments at the array edges, and the positions of rare cutting restriction sites relative to these edges.
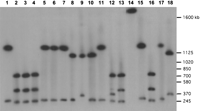
Pulsed-field gel analysis of the DXZ1 array in LT690. High molecular weight DNA was digested with one or two restriction enzymes, separated by electrophoresis, and probed with a DXZ1-specific probe. Fragment sizes were used to derive the restriction map shown in Fig 1A. Enzymes used for each digestion are BglI (lane 1);BglI/BglII (lane 2);BglII (lane 3);KpnI/BglII (lane 4); KpnI (lane 5); BglI/KpnI (lane6); BglI (lane 7);BglI/ScaI (lane 8); ScaI (lane 9); ScaI/KpnI (lane 10);KpnI (lane 11); ScaI/BglII (lane 12); BglII (lane 13);ApaI (lane 14); ApaI/BglI (lane 15); ApaI/BglII (lane16); ApaI/KpnI (lane 17);ApaI/ScaI (lane 18). The image is composed of two halves of the same gel, electronically spliced between lanes13 and 14. Selected S. cerevisiae size markers are shown at the right.
DNAs were digested with ApaI, BglI, BglII,BstEII, KpnI, and ScaI, enzymes that do not cut within the basic repeat sequence of α-satellite DNA (locus DXZ1; Waye and Willard 1985; Mahtani and Willard 1990). Restriction enzyme digests were fractionated by pulsed-field gel electrophoresis and filter transfers were probed with a DXZ1-specific probe (Waye and Willard 1985; Warburton et al. 1991) under high stringency hybridization conditions. Within each α-satellite array, all sites for these restriction enzymes were mapped relative to one another by carrying out double-digests in all pairwise combinations. On either chromosome, single digests with each of these restriction enzymes cut the array into between one and six fragments that hybridized to DXZ1 probe.
DXZ1 array sizes were measured for each enzyme by adding together, for a given digest, all fragment sizes containing α-satellite DNA. Size estimates obtained with each of the enzymes BglI,BglII, BstEII, KpnI, or ScaI were in close agreement (see Fig. 1) and were consistent with results of double digests (see Fig. 2), allowing us to construct restriction maps of each array. The clustering of these restriction sites at the outside edges of the array (Fig. 1) suggests that sequences adjacent to α satellite DNA on either chromosome arm have sequence properties similar to typical single-copy DNA (Tyler-Smith and Willard 1993).
Digestion with ApaI, an enzyme that cuts relatively frequently in typical genomic DNA, released each of the two arrays onto one large fragment ∼200 kb larger than the predicted total array size (Table1). Therefore, whereas the edges of the DXZ1 array are characterized by a clustering of many common-cutting sites, this flanking DNA may not have a typical ApaI restriction site distribution. It is possible that DXZ1 sequences may be flanked on one or both sides by a small amount (∼200 kb or less) of other repetitive sequences such as diverged α satellite (e.g., Cooper et al. 1993; Bayne et al. 1994) or another family of repetitive DNA. In fact, at the centromeres of chromosomes 7 (Wevrick et al. 1992), 8 (Lin et al. 1993), Y (Cooper et al. 1993), and the acrocentric chromosomes (Trowell et al. 1993; Wohr et al. 1996), there is evidence that small amounts of other tandemly repeated DNA families are located near α-satellite sequences.
α-Satellite Array-Containing Restriction Fragment Size Estimates
Whereas the cluster of BglI, KpnI, BstEII, and BglII sites at position 1220 kb in LT690 (Fig. 1) may identify a small amount of DNA with typical restriction site composition at this position in the array (perhaps reflecting small amounts of interspersed, retroposed repetitive elements such as L1; e.g., Wevrick et al. 1992), there is no evidence within either LT690 or AHA–11aB1 for a large (>100 kb) amount of nonrepetitive DNA either interspersed with or present as a large block in X chromosome α-satellite sequences. Such internal fragments containing nonrepetitive (i.e., unique sequence) DNA should not hybridize to the α-satellite probe and would be expected to yield inconsistent size estimates for different enzyme digests. The consistent agreement between array length estimates derived from multiple independent digests with different restriction enzymes suggests that such unique sequence DNA is largely absent.
Mapping the Arrays with Rare-Cutting Enzymes
Whereas common-cutting enzymes digest away most of the nonrepetitive, typical genomic sequences on a given α-satellite-containing fragment, rare-cutting enzymes generally release the entire array along with variable amounts of flanking DNA (depending on the enzyme) onto one very large, often several megabase-sized fragment. Without exception, all conventional rare-cutting enzymes tested with a CpG dinucleotide in their recognition sequence did not cut within the DXZ1 arrays in the two cell lines examined here. For each of the two cell lines, the sizes of theseNarI, SmaI, NruI, MluI, andEagI fragments hybridizing to DXZ1 probe are given in Table 1.
Table 1 also compares for each enzyme, the amount of nonrepetitive, flanking DNA determined to be present on a given α-satellite-containing restriction fragment. This quantity of flanking DNA was estimated by subtracting the size of the α-satellite array from each fragment length. The positions of rare-cutting enzyme sites relative to the edges of the α satellite array on the two X chromosomes (in AHA–11aB1 and LT690) are strongly correlated with one another. For example, in both cell lines, the enzymes ApaI and NarI release fragments ∼200-kb larger than the α satellite array; SmaI cuts ∼400 kb outside the LT690 array and ∼350 kb outside the AHA–11aB1 array. Similarly, in LT690, the NruI fragment is predicted to contain ∼600 kb of single-copy sequence relative to ∼500 kb in AHA–11aB1. In one case, notably MluI, the correlation may be less obvious because of the difficulty, particularly in AHA–11aB1, of accurately sizing very large (>3000 kb) fragments.
On the X chromosome, with the exception of the small array in LT690, the sizes of α-satellite arrays vary continuously from 2200 to 3730 kb as a result of array length polymorphism (e.g., Mahtani and Willard 1990). If restriction sites in the DNA flanking the array are positioned at fixed, predictable distances from the array edges, then, for a given enzyme, the sizes of rare-cutting fragments should follow the same distribution as that of the array lengths itself. Under this hypothesis, the amount of flanking DNA on a restriction fragment that also contains the entire DXZ1 array should be identical for many different X chromosomes digested with the same restriction enzyme. As is shown in Table 1, data from these two extensively mapped arrays support this hypothesis. Thus, the variation in restriction fragment lengths caused by array size polymorphism (Wevrick and Willard 1989;Mahtani and Willard 1990) may taper off close to the edges of the α-satellite array, as this DNA becomes typical euchromatic sequence. More data that precisely position single-copy markers relative to the array edges will help evaluate the validity of this hypothesis. To date, in situ hybridization experiments have failed to identify large, variable blocks of other repetitive DNA families adjacent to DXZ1 sequences (Miller et al. 1995; V. Powers and H.F. Willard, unpubl.), even though such sequences are readily detected at many other centromeres. This also supports the idea that the α-satellite array measured here may lie close to the unique, euchromatic sequences of the two chromosome arms. As suggested by theApaI digests, if families of other repetitive DNAs are present in the vicinity of the centromere of the X chromosome, then these sequences must exist in relatively small (<200-kb) blocks.
When restriction fragments containing α-satellite sequences were tested for cohybridization with probes from the proximal unique sequences DXS422 (in Xp11.21) and DXS62 (in Xq11), we were unable to detect any colocalization with filter hybridization (M.M. Mahtani, data not shown). In LT690, because DXZ1 is found on a single NruI fragment that also contains ∼600 kb of non-α-satellite sequences, the DXS422 and DXS62 loci must lie at least 2100 kb apart and thus 300–600 kb outside the α-satellite array.
Because the small LT690 DXZ1 array is almost exactly half the size of the mean array length from the population sampled, we considered the possibility that the measured array might be present in duplicate on that chromosome. If the LT690 array were itself tandemly duplicated in such a manner, however, then digests with restriction enzymes that cut within the array or very close to its edges (i.e., BglI,KpuI, BglII, etc.) would yield bands of double intensity. Whereas such doublets might not be easily scored in LT690, they should be readily apparent in the DNA of the mother of LT690, who carries both an average-sized array and the small array. Comparisons of the α-satellite gel patterns from the two X chromosomes in the mother, however, revealed no obvious intensity differences between fragments corresponding to one or the other haplotype (M.M. Mahtani, data not shown). Thus, it is unlikely that a simple tandem duplication of only DXZ1 is present in LT690, and we conclude that the actual DXZ1 array size is ∼1500 kb.
Genetic Distance Across the Centromere
Some information is lost when genetic distances are derived from linkage analysis, because only those meioses informative for the relevant markers are considered in the analysis. To examine each chromosome in the CEPH database (i.e., not only those informative for a given pair of markers), instead we constructed 27-locus meiotic maps of inherited X chromosomes from the entire youngest generation (349 female meioses; see Methods) of the CEPH family panel. From this 27-locus marker set, on average 10.1 markers were informative within a given family (range 5–15 loci per family). Because these loci all map to a ∼20-cM region spanning the centromere (Nelson et al. 1995; Weeks et al. 1995), the average intermarker distance is ∼2 cM.
For each of the meioses in the 40 family CEPH panel, positions of recombination events were plotted relative to the inherited maternal X chromosome (both by hand and with the CRIMAP program CHROMPIC) for the 27-marker loci. By use of distal markers to establish phase and to rule out double crossovers in flanking regions, chromosomes were identified that showed evidence for recombination between DXS255 in Xp11.22 and DXS153 in Xq13 (∼15 Mb physical distance; Nelson et al. 1995;Nagaraja et al. 1997). In the data set of 349 meioses, only 26 crossovers occurred in this interval; no double crossovers were detected.
Crossing-over occurs rarely across the centromere, but when it does occur, it seems to cluster in a few families. Specifically, most families showed no evidence for crossing-over in the pericentromeric region, whereas more pedigrees than expected (χ2 = 7.2; P = 0.03) had two or three individuals with single crossovers occurring somewhere between DXS255 and DXS153. This may reflect an underlying genetic (interfamilial) variation in recombination rates.
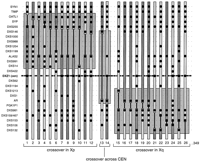
Figure 3 illustrates the inferred position of recombination events on each of the 26 chromosomes in the CEPH panel in which we found evidence for a proximal Xp or Xq exchange. For example, crossovers occurred between loci in proximal Xp in individuals 1–12 and in proximal Xq in individuals 15–26; on the basis of the established maps of the region, each crossover clearly occurred outside of the DXZ1 array and its proximal flanking sequences. Only individuals 13 and 14 showed evidence for meiotic exchanges either within the most centromere proximal intervals or possibly within the centromere itself; however, whereas all available markers were tested in all the relevant meioses, we were unable to more precisely localize the endpoints of these two recombination events because of lack of heterozygosity for the relevant markers. For example, in individual 13, a crossover occurred between DXS146 and AR (Fig. 3). Because this individual was uninformative at the intervening loci, we were unable to further refine the crossover position. Given the large physical distances involved between these loci (Nelson et al. 1995), however, it is likely that these two crossovers actually occurred well outside the centric heterochromatin (see Discussion). Because all but two of the crossovers could be mapped clearly on one side of the centromere or the other (i.e., distal to DXS14 in Xp11.21 or distal to DXS1213 in Xq11), these two markers define the most proximal centromere-spanning region, within which a maximum of two crossovers occurred (Fig. 3). Thus, the maximum genetic distance between these loci can be calculated as 2 recombinants/349 meioses, or 0.57 cM (0 cM–1.4 cM, 95% C.L.). Depending on precisely where these two crossovers actually occurred, the distance across the centromere itself must be <0.57 cM.
Inheritance of loci in the pericentromeric region of the X chromosome in the 26 individuals (of all 349 CEPH meioses; see Methods) in which a recombination event occurred between markers DXS255, DXS153, or intervening loci. Positions of each of the 27 loci are indicated by regularly spaced broken lines and the centromere by bold broken lines (center), and do not reflect genetic distances. Order of these loci is taken from published consensus maps (Nelson et al. 1995; Miller et al. 1995; Nagaraja et al. 1997). Light- and dark-shaded regions, respectively, represent paternally or maternally derived X chromosome material. Between them, solid, vertical lines illustrate regions within which a meiotic recombination event must have occurred. (•) Loci that were both informative and genotyped in a specific meiosis. Recombination events occurred on chromosomes passed to individuals1–12 in Xp, individuals 15–26 in Xq, and possibly across the centromere only in individuals 13 and 14.
DISCUSSION
The convergence of high-density physical and genetic maps of the human genome (Hudson et al. 1995) will allow recombination rates to be directly compared with known physical distances at regular intervals along each of the human chromosomes. Preliminary comparisons suggest that the ratios of physical to genetic map lengths will vary greatly relative to the genomic average. This variation in recombination frequency reflects differences within a chromosome, between chromosomes, and between the sexes. For example, the genetic map is 1.6-fold longer in females than males (Dib et al. 1996), revealing a generally higher rate of exchange in female than male meiosis. Averaged over the genome, the mean rate of exchange is 1 cM every 717 and 1155 kb in females and males, respectively, with a similar range of variation for individual chromosomes (Morton 1991; Dib et al. 1996).
Such chromosome- and genome-averages, however, fail to consider another level of map length variation: the probable abundance of local hot and cold spots. In fact, when long-range (restriction or YAC contig) physical maps are compared with their corresponding genetic maps, large regional variations in crossing-over rates are observed (e.g., Abbs et al. 1990; Allitto et al. 1991; Nagaraja et al. 1997). Unusually high or infrequent recombination rates are thought to be dependent on proximity to local recombination signals, changes in higher order chromatin structure, or presence of chromosomal structural elements such as centromeres or telomeres.
In Drosophila (Dobzhansky 1930; Mather 1939), yeast (Clarke and Carbon 1980), and Neurospora (Davis et al. 1994), recombination at centromeres is markedly reduced relative to that found in other chromosome regions. This repression of recombination persists in Drosophila when adjacent heterochromatin is deleted (Yamamoto and Miklos 1978) and is generated de novo in yeast when a centromere is integrated into a new site on the chromosome (Nakaseko et al. 1986). In humans, chiasmata are found infrequently near chromosome centromeres (Hulten 1974; Hulten et al. 1982), but definitive studies of recombination rates across measured physical distances at human centromeres have only become available recently.
Perhaps most relevant is a comparison of physical distance across the centromere of chromosome 10 with the genetic distance between two centromere-flanking microsatellite markers; this study reported a 10-fold repression of recombination (Jackson et al. 1996), consistent with earlier studies on this chromosome (Carson and Simpson 1991;Lichter et al. 1993). On the other hand, estimates of recombination rates at the pericentromeric regions of chromosomes 11 (Janson et al. 1991) and 21 (Jabs et al. 1991; Van Hul et al. 1993) seem less consistent with a centromeric suppression of meiotic exchange. These apparently conflicting data, however, may be reconciled by the fact that the chromosome 11 map may be bounded by loci a significant distance away from α-satellite sequences, because the physical distance between the centromere and the loci studied is not known. In fact, radiation hybrid mapping data on chromosome 11 suggest that the most proximal centromeric markers span a large physical distance (in centiRads) but exhibit no recombination (James et al. 1994). On chromosome 21, the extensive homology between α-satellite arrays on chromosomes 21 and 13 (for review, see Trowell et al. 1993) may generate ambiguous genotypes of the α-satellite polymorphisms within these arrays, artificially inflating observed recombination rates. Thus, when more definitive data are available, recombination frequencies at the centromeres of these and other chromosomes may be consistent with the repressed rates of meiotic exchange found on the X chromosome (as reported here; see also Fain et al. 1995; Weeks et al. 1995) and chromosome 10 (Carson and Simpson 1991; Lichter et al. 1993;Jackson et al. 1996).
To determine the rate of exchange across the X chromosome centromere and characterize the extent of repression of meiotic exchange in humans, we measured the frequency of genetic exchange across the X chromosome centromere by comparing a long-range restriction map with detailed genetic maps of the CEPH meioses. The physical map across the centromere encompasses on average ∼3000 kb of α-satellite DNA and ∼600 kb of unique sequence DNA, although still exclusive of either of the most centromere-proximal genetic markers, DXS422 (ZXDA) or DXS62 (Miller et al. 1995; Nelson et al. 1995). The maximum genetic distance across this region was evaluated by identifying positions of recombination events relative to 27 loci in the pericentromeric region of the X chromosome. At most, two chromosomes from a sample of 349 female meioses of the CEPH family panel were recombined in the genetic intervals spanning or including the centromere (Fig. 3). Therefore, the maximum genetic distance across the 3600-kb region is ∼0.57 cM, suggesting an upper limit on the rate of exchange in this region of 1 cM/6300 kb (i.e., 0.57 cM/3600 kb).
This recombination frequency is much lower than that expected from a similar-sized interval elsewhere on the X chromosome. For example, assuming an average frequency of exchange of ∼1 cM/800 kb (the genetic to physical distance ratio when averaged over the entire X chromosome; Dib et al. 1996; Nagaraja et al. 1997), a 3600-kb interval elsewhere on this chromosome would have an expected map length of 4.6 cM (2.4 cM–6.8 cM, 95% C.L.). In a sample size similar to that examined here, over a genetic distance of 4.6 cM, one would expect to observe about 16 crossovers. Our observation of only 2 crossovers is well outside the predicted Poisson variation around this mean (P = 1.4 × 10−5), indicating a highly depressed rate of exchange at the centromere relative to the chromosome average. This repression provides initial quantitation of a more general effect inferred on the proximal X chromosome, when genetic distances are compared with rough cytogenetic band sizes (Fain et al. 1995; Weeks et al. 1995) or to a YAC contig map (Nagaraja et al. 1997).
The calculated genetic distance of 0.57 cM is a conservative estimate of the extent of centromeric repression, because it reflects the genetic map length of a region that includes, but is not limited to, the ∼3600 kb mapped here. Therefore, the rate of exchange reported here overestimates crossing over unless both recombination events fall within the most proximal 3600 kb, including the α-satellite array. In fact, this genetic distance of 0.57 cM may fall in any interval, or be evenly distributed, within the centromere-spanning region. The consensus map of the X chromosome indicates that this region may encompass up to 8–10 Mb of DNA, including the length of the α-satellite array (Nelson et al. 1995). Once the gaps in the physical map of the X chromosome are closed, a definitive rate of exchange can be calculated for the entire interval. Because it is possible that the genetic distance of 0.57 cM may be distributed over as much as 10,000 kb, the recombination rate across the centromere (including centromere adjacent DNA), may be as low as 1 cM/17,000 kb. Nonetheless, this rate should be taken as an upper limit for the rate of crossing over at the centromere itself, keeping in mind that it is formally possible that, as in Drosophila (Baker 1958), recombination may not occur at all in proximal heterochromatin.
Regional variation in recombination rates on the X chromosome is extensive. In fact, when long-range physical maps are compared with their genetic counterparts, different regions of the X chromosome have widely different recombination rates. For example, 1 cM corresponds to 190 kb in the DMD region (van Ommen et al. 1986; Abbs et al. 1990), 340–800 kb near the end of the long arm [Xq27.2-qter (Kenwrick and Gitschier 1989; Poustka et al. 1991)], 600 kb near the end of the short arm [Xp22.3-pter (Petit et al. 1990)], 640 kb in Xq26 (Little et al. 1992), and >5 Mb in Xq13.3-q21.3 (Nagaraja et al. 1997). Whereas recombination rates may be reported more frequently for regions of high recombination, recombination rates approaching that reported here for the centromere have not been documented for other regions on the X or, until recently (Jackson et al. 1996), for other regions of the genome.
METHODS
Physical Mapping
To generate restriction maps of the DXZ1 α-satellite array on the X chromosome, we chose restriction enzymes that typically cut mammalian DNA into small (<20 kb) fragments, but that fail to cut within the basic 2.0-kb higher-order repeat sequence on the X chromosome (Waye and Willard 1985; Mahtani and Willard 1990). High molecular weight DNA was prepared and digested as described (van Ommen and Verkerk 1986; Wevrick and Willard 1989). When restriction buffers were incompatible between two enzymes, double digests (of DNA in agarose blocks) were done sequentially for 4 hr each, separated by a 1-hr rinse in 10 mm Tris-Cl; 10 mm EDTA. S. cervisiae and S. pombe chromosomes (Bio-Rad) were used as high molecular weight size standards. The gel-purified PCR product of X chromosome specific α-satellite primers X-3A and X-4A (Warburton et al. 1991) or the plasmid pBamX7 (Waye and Willard 1985) was used as a probe for DXZ1. Pulsed-field (CHEF) gel electrophoresis, Southern blotting, and hybridization were performed as described (Waye et al. 1988; Wevrick and Willard 1989; Mahtani and Willard 1990).
LT690 is a lymphoblastoid cell line derived from a normal Atlantic Canadian male shown previously to carry a particularly small X chromosome α-satellite array (Mahtani and Willard 1990). AHA-11aB1 is a mouse/human somatic cell hybrid containing an X chromosome as its only human material (Willard 1983).
Genetic Mapping
DNA from the 40 family panel of the CEPH was used in genetic mapping studies. This panel contains 362 female meioses in the youngest generation of primarily three-generation families; therefore, most of these were phase-known meioses. Exactly 349 of these individuals were informative for enough markers in this region such that gaps of no greater than 6 cM existed between adjacent informative loci; the remaining 13 meioses were removed from the calculation of recombination rates to eliminate the possibility of undetected double-crossovers in those regions.
Genotypic data from 27 loci in the pericentromeric region of the X chromosome were compiled from a previously published 18-point linkage map (Mahtani et al. 1991) and studies of microsatellites in proximal Xq (Mahtani and Willard 1993), the ALAS2 gene (Cox et al. 1992), the ZXDA gene (locus DXS422; G. Greig and M.M. Mahtani, data not shown), and the CEPH database V7.1 to generate meiotic maps (Fain et al. 1995). All available individuals in CEPH were genotyped for at least 18 of the loci (Mahtani et al. 1991). Those with pericentromeric crossovers (Fig.3) were then typed for all additional loci. The total of 27 loci span a genetic distance of ∼20 cM in the pericentromeric region (Mahtani et al. 1991; Fain et al. 1995; Weeks et al. 1995), extending from the cytogenetic band Xp11.22 on the short arm, across the centromere and into band Xq12 on the long arm (Miller et al. 1995; Nelson et al. 1995). Genotypes for markers DXS7 in Xp11.3 and PGK1 in Xq13.3, two flanking loci that map distal to the region examined here, were used to ensure consistency and to eliminate genotyping errors from the dataset. Locus order was determined as described previously (Mahtani et al. 1991) and is consistent with consensus maps of the X chromosome (Wang et al. 1994; Fain et al. 1995; Nelson et al. 1995; Weeks et al. 1995).
Acknowledgments
We thank David Botstein, Rick Myers and Luca Cavalli-Sforza for support; and David Botstein, Pat Brown, David Cox, Eric Lander, and Andrew Kirby for their interest and comments. This work was supported by National Institutes of Health grant HG00107 to H.F.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.