Snapshot of a Large Dynamic Replicon in a Halophilic Archaeon: Megaplasmid or Minichromosome?
Abstract
Extremely halophilic archaea, which flourish in hypersaline environments, are known to contain a variety of large dynamic replicons. Previously, the analysis of one such replicon, pNRC100, inHalobacterium sp. strain NRC-1, showed that it undergoes high-frequency insertion sequence (IS) element-mediated insertions and deletions, as well as inversions via recombination between 39-kb-long inverted repeats (IRs). Now, the complete sequencing of pNRC100, a 191,346-bp circle, has shown the presence of 27 IS elements representing eight families. A total of 176 ORFs or likely genes of 850-bp average size were found, 39 of which were repeated within the large IRs. More than one-half of the ORFs are likely to represent novel genes that have no known homologs in the databases. Among ORFs with previously characterized homologs, three different copies of putative plasmid replication and four copies of partitioning genes were found, suggesting that pNRC100 evolved from IS element-mediated fusions of several smaller plasmids. Consistent with this idea, putative genes typically found on plasmids, including those encoding a restriction-modification system and arsenic resistance, as well as buoyant gas-filled vesicles and a two-component regulatory system, were found on pNRC100. However, additional putative genes not expected on an extrachromosomal element, such as those encoding an electron transport chain cytochrome d oxidase, DNA nucleotide synthesis enzymes thioredoxin and thioredoxin reductase, and eukaryotic-like TATA-binding protein transcription factors and a chromosomal replication initiator protein were also found. A multi-step IS element-mediated process is proposed to account for the acquisition of these chromosomal genes. The finding of essential genes on pNRC100 and its property of resistance to curing suggest that this replicon may be evolving into a new chromosome.
[The sequence data described in this paper have been submitted to GenBank under accession no. AF016485.]
Both archaeal and bacterial prokaryotes generally contain multiple circular replicons from only a few kilobases to several megabases in their genomes. Since early genetic work onEscherichia coli established the dogma of a single circular chromosome in prokaryotes, essentially all smaller replicons in cells have been relegated to the status of extrachromosomal elements or plasmids, irrespective of the species (Drlica and Riley 1990). Of the many plasmids that have been characterized, several are megaplasmids from a hundred kilobases to megabase sizes, including F (fertility) and R (resistance) factors, and toxin-bearing plasmids in E. coliand other pathogenic bacteria (Proter 1991), tumor-inducing plasmids and symbiotic plasmids in Agrobacterium and Rhizobiumspecies (Van Larebeke et al. 1974; Banfalvi et al. 1985), and a variety of aromatic hydrocarbon degradation plasmids in Pseudomonasspecies and related bacteria (Franz and Chakrabarty 1986; Choudhary et al. 1997; Mouncey et al. 1997). Over the last few years, many of these and other large replicons have been studied and some found to contain genes that are normally thought to be essential for cell viability. The most striking example is for Rhodobacter spheroides, which contains a 900-kb replicon with two copies of the rRNA operon and a variety of other essential genes (Suwanto and Kaplan 1989; Choudhary et al. 1997). This finding led to the suggestion that prokaryotic genomes may be composed of multiple essential replicons and opened the possibility that some replicons originally classified as megaplasmids may in fact be chromosomes (Allardet-Servent et al. 1993; Michaux et al. 1993; Zuerner et al. 1993; Cheng and Lessie 1994). However, chromosomal status may depend not just on the occurrence of essential genes, but also on other criteria such as size, copy number and replication control, and evolutionary history.
The recent advent of high throughput sequencing technology has provided an outstanding opportunity for detailed analysis of prokaryotic genomes (Fleischmann et al. 1995; Fraser et al. 1995, 1997; Bult et al. 1996;Himmelreich et al. 1996; Kaneko et al. 1996; Sensen et al. 1996;Blattner et al. 1997; Klenk et al. 1997; Kunst et al. 1997; Smith et al. 1997; Deckert et al. 1998; Cole et al. 1998). In addition to permitting comparative phylogenetic analysis of individual genes across a wide spectrum of organisms, these studies also promise a better understanding of genome evolution and dynamics, including greater insights into the relationships between replicons resident within individual species. With the evolution of prokaryotic genomes in mind, we have focused on the extremely halophilic archaeon,Halobacterium sp. strain NRC-1, which grows optimally at a nearly saturated (4.5 m) NaCl concentration (Vreeland and Hochstein 1993; DasSarma and Fleischmann 1995). The genome ofHalobacterium NRC-1 is notable for the presence of dynamic replicons and a variety of transposable elements that give rise to frequent DNA rearrangements (Charlebois and Doolittle 1989;DasSarma 1993). The genome of NRC-1 contains a circular 2-Mb chromosome and two other large replicons, pNRC100 and pNRC200 (Hackett et al. 1994). We have focused on pNRC100, a 191-kb replicon, which was restriction mapped and shown to contain at least 17 insertion sequence (IS) elements and large (>35 kb) inverted repeats mediating inversion isomerization (Ng et al. 1991). pNRC100 was also shown to contain a cluster of genes specifying buoyant intracellular gas-filled vesicles (Halladay et al. 1992, 1993; DasSarma et al. 1994). Because gas vesicles are necessary for flotation, which enhances both aerobic respiration and photophosphorylation, the finding of these important genes on an extrachromosomal element was surprising.
Here, we report the complete nucleotide sequence of pNRC100 and analysis of its IS elements and coding capacity. We also discuss a hypothetical model for its evolution and possible justification for its status as a minichromosome.
RESULTS
pNRC100 Sequencing and Assembly
Sequencing was conducted on shotgun libraries ofHalobacterium sp. strain NRC-1 plasmid DNA and on cloned and ordered HindIII fragments of pNRC100. The initial period of automated sequence assembly produced nine contigs representing pNRC100 ranging from 1 to 69 kb. The assembly process was challenging because of the presence of a total of 27 copies of IS elements (17% of the replicon; Table 1) and the large (∼39 kb) inverted repeat (IR). The contigs containing two or more physically unlinked segments of pNRC100 sequence were resolved by comparison of regions of IS element heterogeneities (e.g., for ISH3, ISH7, and ISH8; Charlebois and Doolittle 1989; DasSarma 1993) and alignment of known target-site duplications flanking IS elements (Ng et al. 1991) manually by use of the FINDPATTERNS, BESTFIT, SEQED, and WORDSEARCH programs in the GCG software package (Devereux et al. 1984). End sequencing of the ordered pNRC100 HindIII fragment library (Ng et al. 1991) provided a scaffold for assembly. Appropriate segments of chimeric contigs were dissected and reassembled into a circular 191,346-bp sequence (GenBank Accession no. AF016485).
IS Elements in pNRC100
pNRC100 Putative Genes and Gene Products
A total of 1,965 individual ORFs 15 bp or larger were identified and analyzed by determination of their size, location, GC composition, codon third position GC bias, and isoelectric point of the predicted protein product by use of the GCG software package. These criteria were used to select 176 probable genes, 39 of which were repeated in the large IR (Fig. 1). Because one H0761 copy on the IR is interrupted by an ISH2 element, resulting in two smaller ORFs, the number of different ORFs is 136. Our analysis indicated that 72% of the replicon is coding and 28% is noncoding.
Linear representation of the genes, ORFs, and IS elements of the circular plasmid pNRC100. The plasmid is divided into five sections, the small single-copy region, the right IR (red arrow), the large single-copy region (divided into two sections), and the left IR (red arrow). IS elements (arrows indicating orientation) are shown along with a scale in nucleotides and genes and ORFs on each DNA strand are shown below. Identities and orientation of genes (red or green arrows) and ORFs (purple or blue arrows) are indicated. All 1965 pNRC100 ORFs five codons or larger have been assigned numerical designations on the basis of location; however, only those ORFs judged to represent probable genes are shown for clarity. Additional information is available at the following Web site:http://chroma.mbt.washington.edu/seq_www/.
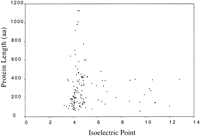
Of the 136 different probable gene products encoded by pNRC100, 62 or about 46% had statistically significant hits to the databases (Table2). Of these, 14 hits were to gas vesicle proteins, 8 were to replication or partitioning proteins, 7 were to regulatory proteins, 4 were to transcription factors, 4 were to membrane components, 2 were to redox proteins, 2 were to heavy metal resistance proteins, 2 were to DNA endonucleases, and 1 was to a helicase. The remaining 18 hits were to putative transposases encoded by IS elements or proteins with unknown functions. Of the closest matches, 36 were to archaeal proteins, 23 were to bacterial proteins, and 3 were to eukaryotic proteins. Thus, like other archaea,Halobacterium clearly has both bacterial features, such as metabolic proteins, and eukaryotic features, such as the transcription system (Koonin et al. 1997). An additional interesting finding was that like most characterized proteins of halophiles, the great majority (91%) of the pNRC100 probable gene products are acidic (Fig.2), a feature likely to be important for function at high salt concentrations known to be present in their cytoplasm (Lanyi 1974; Dym et al. 1995).
Summary of 62 Probable pNRC100 Gene Products with Homologs in GenPeptides Database
Plot of calculated isoelectric point versus length of 136 predicted proteins encoded by pNRC100. Isoelectric points were calculated by use of the GCG isoelectric program (Devereux et al. 1984). The range of calculated pI was 3.24–12.81 and the average pI was 5.05. The range of sizes was 56–1128 amino acids and the average size was 290 amino acids.
Among the most interesting putative gene products encoded by pNRC100 were the following: (1) H0378, H0486, H1211, and H1356 are probably transcription factors similar to the eukaryotic and archaeal general transcription factor TBP (TATA-binding protein; Thomm 1996). The multiplicity of TBPs suggested by this finding is highly unusual. However, not all of them may be functional. For example, H0378 appears to be only one-half the size of the others. (2) H0761, H0136, H1080, and H1260 are similar to replication proteins. H0761 is in the Orc1p family of eukaryotic origin-binding proteins (Gavin et al. 1995) whereas the other three are members of a family of replication proteins in halophiles (Ng and DasSarma 1993). The gene encoding H0136,repH, was shown previously to be required for replication of pNRC100 minireplicons (Ng and DasSarma 1993). (3) H0066, H0324, H0722, and H0991 are similar to the ParA family in bacteria and to Soj inBacillus subtilis and Methanococcus jannaschii, which are involved in plasmid and chromosomal partitioning, for orderly segregation of replicated molecules into daughter cells (Wheeler and Shapiro 1997). (4) H0511 and H0520 are similar to subunits I and II ofE. coli cytochrome d oxidase, a terminal electron acceptor in the electron transport chain (Miller and Gennis 1983). (5) H0606 and H0610 are similar to thioredoxin and thioredoxin reductase, which are used for reduction of ribonucleotides to deoxyribonucleotides for DNA synthesis (Neuhard and Nygaard 1987). (6) H0337 is similar to several protein kinases of two-component regulatory systems, including both components of the FixL/J system probably involved in oxygen regulation (David et al. 1988; Lois et al. 1993). (7) H1450 and H1477 are similar to arsenic-resistance proteins, with the former similar to the catalytic subunit of the arsenite pump, and the latter similar to arsenate reductase (Rosen et al. 1988, 1991). (8) H1161 is similar to the type IV restriction-modification system protein Eco57I which contains both restriction and modification activity (Janulaitis et al. 1992). (9) H1537 is similar to an intron encoded endonuclease (Turmel et al. 1995). (10) Over a dozen Gvps are thought to be involved in gas-vesicle formation and have been studied genetically (Halladay et al. 1992, 1993; DasSarma et al. 1994).
DISCUSSION
The probable genes on pNRC100 with statistically significant homologs in the databases fall into three categories. Some genes appear to be typical for plasmids and extrachromosomal elements: for example, replication and partitioning genes, and arsenic resistance. Other genes, for example the gas vesicle genes and a number of regulatory genes, are not typically extrachromosomal but their extrachromosomal location is not surprising given that they may be dispensable for cell viability. Quite unexpectedly, however, another set of genes appears to encode essential proteins. For example, cydA and cydBencode a cytochrome d oxidase, which is used for the terminal step in respiration, and trxA and trxB encode thioredoxin and thioredoxin reductase enzymes, which participate in the pathway for synthesis of deoxyribonucleotides. These enzymes are involved in fundamental cellular processes and appear to be required for cell viability. Moreover, Southern hybridization analysis showed that these are unique genes present only on pNRC100 and are not repeated elsewhere in the genome (data not shown). Transcription factors, such as TBPs, four of which are encoded by pNRC100 (tbpA, tbpB, tbpC, and tbpD), may also be required under conditions in which they are used for transcription of critical genes. Therefore, pNRC100 appears to be a replicon with unique essential genes expected to reside on a chromosome rather than a plasmid.
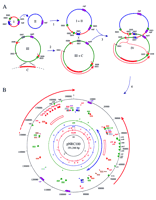
The finding of chromosomal genes on pNRC100 raises the question of the mechanism of acquisition of these genes (Fig. 3). One possibility is that a smaller plasmid had integrated into theHalobacterium chromosome in the past, perhaps mediated by an IS element, and had subsequently excised imprecisely capturing a segment of chromosomal genes in the process. This type of process has been shown to be used to generate F′ plasmids in E. coli(Holloway and Low 1987). The IRs of pNRC100 would be prime candidates for recent acquisition from the chromosome as the cytochrome d oxidase (cydA and cydB) and thioredoxin/ thioredoxin reductase (trxA and trxB) genes, as well as one of the TBP genes (tbpB) are all located there. The presence of a eukaryotic-like chromosomal replication initiator protein gene (H0761) in the IRs is also intriguing in this respect (Gavin et al. 1995). This region also contains a large number of putative genes with homology to genes present on the chromosomes of other microorganisms, for example, the B. subtilis yrkE, yrkF, yrkH, and yrkJ genes, which are of unknown functions (Kunst et al. 1997), and HI1604, a Haemophilus influenzae putative phosphate transport gene (Fleischmann et al. 1995). In addition to bearing what appear to be chromosomal genes, the IR region is also relatively GC rich (Fig. 3) and IS-element poor, both characteristics of theHalobacterium chromosome (Charlebois and Doolittle 1989;DasSarma 1993; Hackett et al. 1994). An additional interesting possibility is that the inverted duplication of genes in the IR region may be important for stability. Duplicated genes present in inverted orientation are less likely to be lost by deletion and inactivated by deleterious mutation, as a result of repair by copy choice mechanisms. Analogous functions have been proposed for the large IRs in several chloroplast chromosomes, which are of similar size and arrangement to pNRC100 (Palmer 1985).
(A) Hypothetical pathway for evolution of pNRC100 (not drawn to scale). Step 1: Two small plasmids (I + II) fused, probably mediated by an ISH3 element, to form a larger plasmid (I + II) representing most of the large single-copy region of pNRC100. Step 2: A region of the chromosome (C, thick red line), 15 kb or larger was acquired on a plasmid (III) by integration into and aberrant excision out of the chromosome. The acquired chromosomal region became associated with a 50-kb composite transposon bounded by IRs of ISH8 on a large plasmid (III + C) containing one copy of the 39-kb IR region (red arrow) and most of the small single-copy region of pNRC100. Step 3: The intermediate plasmids fused, probably through ISH7-mediated homologous or site-specific recombination. Step 4: The IR region of pNRC100 was duplicated by a gene conversion-like mechanism (indicated by large orange bracket with arrow) involving ISH2 and ISH3 elements at the ends of the IRs and between the small and large single-copy regions. Subsequent insertion of an ISH2 and ISH3 element into one copy of the IRs is not shown.The relative locations of rep genes, IS elements, and the acquired chromosomal region are indicated by colored boxes. Heterogeneity of the two ISH7 copies is indicated by shading. Target-site duplications are indicated by asterisks. (B) Circular representation of general chromosomal features of pNRC100. The large IRs are indicated on the outside by red arrows and the scale is indicated on the outer circle in nucleotides. The position and orientation of genes are indicated by wide colored arrows. The locations of 15-kb GC-rich (64% G+C) regions within the IRs are indicated by open rectangles. The concentric circles near the center indicate the SfiI, HindIII, DraI, andAflII restriction maps. Restriction fragments are labeled according to size. IS elements flanked by target-site duplication sequences are underlined. Only the genes and ORFs involved in duplicated plasmid features are indicated. (Two-headed blue arrow) Location of a putative 50-kb transposon.
On the basis of the arrangement of IS elements and genes on pNRC100, we have hypothesized the pathway for evolution of the replicon. The original events leading to formation of the large IRs in pNRC100 probably involved multiple IS element-mediated rearrangements. The present location of one copy of the IRs may be explained by the transposition of a ∼50 kb composite transposon. This putative transposon is bounded by IRs of ISH8, which are flanked by short direct repeats (5′-CGTATCGGAG-3′), suggestive of a recent transposition event (asterisks and double arrows in Fig. 3B). Both copies of the IRs are bounded by two other IS elements, ISH2 and ISH3, which were probably involved in the duplication events resulting in formation of the IRs. A possible mechanism for duplication is via gene conversion (or gap repair) of sequences located between the ISH2 and ISH3 elements flanking the original copy of the IRs to another pair of elements located between the small and large single-copy regions (step 4, Fig. 3). Either site-specific or homologous recombination within ISH3 and ISH2 element pairs may have initiated the conversion process, involving either a single molecule or sister plasmids. Such a mechanism is consistent with the finding of short direct repeats flanking one copy of the terminal ISH2 and ISH3 elements but not the other copy of the elements (double asterisks in Fig. 3B). After duplication, two additional transposition events also occurred, insertion of ISH2 and ISH3 elements near one end of one copy of the IR, and resulted in the observed heterogeneity in the IRs. The lack of other sequence heterogeneity between the two copies of IRs and the report of a pNRC100-like plasmid lacking the IRs in another Halobacteriumstrain suggests that the inverted duplication occurred recently in the evolutionary history of NRC-1 (Pfeifer et al. 1981).
The events preceding the acquisition of chromosomal genes and duplication of the IRs in evolution of pNRC100 were likely to be fusions of three smaller plasmids. One replicon fusion was probably mediated by recombination between two nonidentical ISH7 elements present on distinct replicons (I + II and III + C in Fig. 3A). This possibility is suggested by the occurrence of short direct repeats (5′-CGAAGCG-3′) flanking the two copies of ISH7 in pNRC100, which are 85 kb apart, one located to the left of the ISH7 element in the small single-copy region and the second located to the right of the ISH7 element in the large single-copy region (asterisks in Fig. 3A). The smaller of the two replicons hypothesized may have been formed by another replicon fusion, a step suggested by the presence of two very similar replication genes, repI and repJ, in the large single-copy region of pNRC100. Although a specific mechanism is not obvious in this step, the presence of several ISH3 elements in this region suggests their involvement in the fusion process.
The sequence of pNRC100 has provided valuable insights into the structure and evolution of a dynamic replicon in an unstable archaeal genome. A total of 136 probable genes (with 39 duplicated in the inverted repeat) and 27 IS elements have been found. On the basis of the arrangement of IS elements and flanking sequences, we have been able to hypothesize a series of recombinational events that explains the evolution of pNRC100. Several replicon fusions occurred to form a large and complex plasmid. During this process, one plasmid probably inserted into the resident chromosome and excised aberrantly, taking with it a number of genes that were necessary for viability. To stabilize the required chromosomal genes, a 39-kb segment of pNRC100 was duplicated to form IRs, a structure reminiscent of the chloroplast chromosome.
The finding of essential genes on pNRC100 raises an interesting question on the precise distinction between plasmids and chromosomes. Classically, prokaryotic genomes have been thought as being composed of a single large (megabase-plus size) chromosome containing all essential genes, and a diversity of small (<∼100 kb in size) multicopy plasmids containing accessory genes. Plasmids have been shown to recombine with each other or with the chromosome, but are not thought to be involved in the formation of new chromosomes. Not fully consistent with these ideas are pNRC100 and a variety of other essential replicons that have been studied (Van Larebeke et al. 1974;Banfalvi et al. 1985; Franz and Chakrabarty 1986; Suwanto and Kaplan 1989; Allardet-Servent et al. 1993; Michaux et al. 1993; Zuerner et al. 1993; Cheng and Lessie 1994; Choudhary et al. 1997; Mouncey et al. 1997). Many of these seem to occupy an intermediate status between plasmids and chromosomes and may represent evolutionary intermediates in the formation of new chromosomes (or the breakdown of old ones). More detailed analysis of such replicons, including the elucidation of their copy number and replication control as well as their evolutionary history may provide a more meaningful distinction between megaplasmids and minichromosomes. Such scrutiny will likely lead to a deeper understanding of the mechanisms of prokaryotic genome evolution.
METHODS
Plasmid Preparation, Library Construction, and Sequencing
A modified Currier and Nester procedure followed by CsCl–ethidium bromide equilibrium gradient centrifugation was used to purify covalently closed circular DNA from Halobacterium NRC-1 (Ng et al. 1995). A shotgun library was prepared from the purified DNA, which consisted largely of pNRC100, and minor quantities of deletion derivatives of pNRC100, as well as the larger resident plasmid, pNRC200, by sonic shearing and cloning of 1- to 2.5-kb fragments into the SmaI site of M13mp18 (Messing 1983). Sequencing was also conducted at the ends of the cloned HindIII-A to -K fragments of pNRC100 and on shotgun libraries of the pNRC100 HindIII fragments cloned in M13mp18 (Ng et al. 1991). A total of 4,606 sequencing reactions were conducted on pNRC100 subclones by use of fluorescent dye primers or dideoxy-terminators, or both and analyzed on ABI 373 and 377 sequencers (Smith et al. 1986).
Sequence Assembly
The sequencing results were assembled by use of the PHRED/PHRAP/CONSED base-calling and sequence-assembly software (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). The contig sequences were analyzed and merged together by use of FINDPATTERNS, BESTFIT, SEQED, and WORDSEARCH programs in the GCG software package (Devereux et al. 1984).
Sequence Analysis
The complete pNRC100 nucleotide sequence was analyzed for genes by use of a modified GCG FRAMES program (S. Ciufo and S. DasSarma, unpubl.). ORFs 15 bp or larger were identified and analyzed by determining their size, location, GC composition, codon third position GC bias, and isoelectric point of the predicted protein product by use of the GCG software package. The pNRC100 nucleotide sequence and all of the predicted ORF products were used as queries to search for homologous entries in the GenBank and GenPeptide databases with the NCBI BLASTN, BLASTP, and BLASTX programs (Lipman and Pearson 1985). Significance of possible similarities was evaluated by use of the GCG Gap program with multiple randomizations of the query sequence; those homologies with 99% or higher confidence level were included in Table 2.
Acknowledgments
We thank Dr. Samuel Kaplan for critical reading of the manuscript. This work was supported by NSF grants BIR9214821 to L.H. and MCB-9604443 to S.D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵2 Corresponding authors.
-
E-MAIL dassarma{at}microbio.umass.edu; FAX (413) 545-1578; E-MAILleehood{at}u.washington.edu; FAX (206) 616-5197.
-
- Received June 24, 1998.
- Accepted September 21, 1998.
- Cold Spring Harbor Laboratory Press