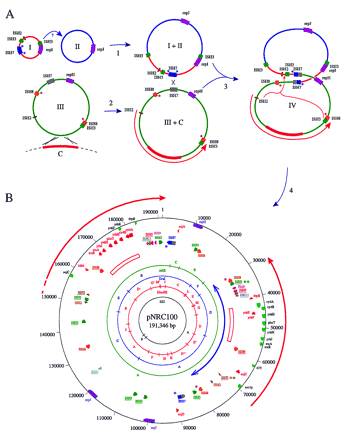
(A) Hypothetical pathway for evolution of pNRC100 (not drawn to scale). Step 1: Two small plasmids (I + II) fused, probably mediated by an ISH3 element, to form a larger plasmid (I + II) representing most of the large single-copy region of pNRC100. Step 2: A region of the chromosome (C, thick red line), 15 kb or larger was acquired on a plasmid (III) by integration into and aberrant excision out of the chromosome. The acquired chromosomal region became associated with a 50-kb composite transposon bounded by IRs of ISH8 on a large plasmid (III + C) containing one copy of the 39-kb IR region (red arrow) and most of the small single-copy region of pNRC100. Step 3: The intermediate plasmids fused, probably through ISH7-mediated homologous or site-specific recombination. Step 4: The IR region of pNRC100 was duplicated by a gene conversion-like mechanism (indicated by large orange bracket with arrow) involving ISH2 and ISH3 elements at the ends of the IRs and between the small and large single-copy regions. Subsequent insertion of an ISH2 and ISH3 element into one copy of the IRs is not shown.The relative locations of rep genes, IS elements, and the acquired chromosomal region are indicated by colored boxes. Heterogeneity of the two ISH7 copies is indicated by shading. Target-site duplications are indicated by asterisks. (B) Circular representation of general chromosomal features of pNRC100. The large IRs are indicated on the outside by red arrows and the scale is indicated on the outer circle in nucleotides. The position and orientation of genes are indicated by wide colored arrows. The locations of 15-kb GC-rich (64% G+C) regions within the IRs are indicated by open rectangles. The concentric circles near the center indicate the SfiI, HindIII, DraI, andAflII restriction maps. Restriction fragments are labeled according to size. IS elements flanked by target-site duplication sequences are underlined. Only the genes and ORFs involved in duplicated plasmid features are indicated. (Two-headed blue arrow) Location of a putative 50-kb transposon.











