Clone–Contig and STS Maps of the Hereditary Hemochromatosis Region on Human Chromosome 6p21.3–p22
Abstract
YAC-based and bacterial-clone based STS-content maps were constructed that served as the framework physical maps for the positional cloning of a candidate gene for hereditary hemochromatosis. The YAC-based map comprises 43 YACs and 86 STS and spans ∼8 Mb of DNA between the class I region of the major histocompatibility complex on human chromosome 6p21.3 and D6S276 in 6p22. Comparison with published maps revealed a hole in the MIT/Whitehead and CEPH YAC maps that includes the immediate region around the hemochromatosis gene itself. Approximately 3 Mb of DNA was covered by a bacterial clone contig that consists of 38 BACs, 45 PACs, 26 P1 clones and one λ phage. The bacterial clone-based STS map comprises 153 STSs. A contiguous block of 8 STSs could be amplified from both human chromosome 6 and 5. Further characterization of selected STSs and bacterial clones by radiation hybrid mapping and fluorescence in situ hybridization, respectively, revealed the presence of a multicopy DNA segment, more than one bacterial clone length in size, which is duplicated near the chromosome-6 centromere and part of which is present in multiple copies on chromosome 5. Possible implications of the incomplete public YAC–contig map and of the multicopy segment for physical mapping and linkage disequilibrium studies of the hemochromatosis candidate region are discussed.
[The sequence data described in this paper have been submitted to GenBank under accession nos. G31205–G31325.]
Hereditary hemochromatosis is one of the most prevalent inherited single-gene disorders in populations of northern European origin (Edwards et al. 1988). The disorder is characterized by the excessive accumulation and deposition of iron in a variety of organs leading to conditions that include arthritis, cardiac dysfunction, cirrhosis, diabetes, and hepatomas (for review, seeMcKusick 1994).
Because of its location near the polymorphic major histocompatibility complex (MHC) on the short arm of chromosome 6, hemochromatosis was among the first human disease loci to be positioned on the human genome map by association and linkage analysis more than 20 years ago, with an increased frequency of the HLA-A3 and HLA-B14 antigens among hemochromatosis patients indicating a genetic founder effect in the natural history of the disease (Simon et al. 1976, 1977; Cartwright et al. 1978).
Recently, a linkage–disequilibrium–mapping approach led to the identification of a gene that is mutated in hemochromatosis patients, >80% of whom are homozygous for a single ancestral mutation (Feder et al. 1996). This gene, HLA-H, is similar to MHC class I-like genes, but is located >3 Mb telomeric of the most distal cluster of previously known class I genes within the MHC. The positional cloning of HLA-H required the detailed analysis of a wide search area encompassing >6 Mb immediately distal to the MHC.
In this report we describe a yeast artificial chromosome (YAC)-based physical map representing the entire candidate region, and a bacterial clone contig spanning ∼3 Mb including the HLA-H gene itself. We discuss some peculiarities of this segment of chromosome 6, and of maps thereof, which may have confounded the search for the hemochromatosis gene. In Ruddy et al. (this issue), expressed sequences identified in the vicinity of HLA-H are described.
RESULTS
YAC–Contig Map
To obtain YAC clone coverage in the segment of human chromosome 6p immediately distal to the MHC, we first screened the Centre d’Etude du Polymorphisme Humain (CEPH) MegaYAC library for the markers D6S265 andHLA-F, located in the class I region at the telomeric end of the MHC (Koller et al. 1989), as well as for markers D6S258, D6S306, D6S105, D6S464, and D6S276, located farther telomeric. Additional YAC clones harboring DNA from this region were detected in the CEPH/Généthon (Chumakov et al. 1995) and MIT/Whitehead (Hudson et al. 1995) databases.
Despite the relatively small genetic distance—all of these markers have been reported to lie within 1 cM of each other (Dib et al. 1996)—the corresponding DNA was initially recovered in three nonoverlapping sets of clones rather than one single contig. Several rounds of bidirectional walking steps using sequence-tagged sites (STSs) developed from clone insert ends were necessary to connect the group of YACs around D6S306, D6S105, and D6S464 with the two flanking contigs containing markers D6S265, HLA-F, D6S258, and D6S276, respectively. Additional STSs were generated, including many polymorphic (CA)n-repeat markers (Feder et al. 1996), and complemented with published STSs. The final collection of 43 YAC clones was tested for the presence or absence of 86 STSs, 36 of which represent YAC insert ends. The resulting STS content map is shown in Figure 1.
YAC-contig and STS map representing an 8-Mb segment of human chromosome 6p21.3–p22 between the distal end of the MHC and D6S276. Forty-three YACs were tested by PCR for the presence or absence of 86 STSs, indicated along the top. STSs in boldface type have not been described previously. STSs marked with a solid circle (•) above the STS name are included in the bacterial clone-based STS content map in Fig. 2. The STSs were ordered so as to minimize the number of YACs with a non contiguous complement of STSs. Groups of STSs not uniquely ordered based on the STS content of the YACs are indicated by solid bars below the STS names. Triple bars indicate adjacent STS pairs connected by less than two independent YACs. YACs are named by the prefix y followed by their respective library address. YACs are depicted as shaded horizontal bars, whose lengths represent their STS content rather than their physical length. YAC sizes (in Mb) are indicated in the second column. Columns 3 and 4 contain information regarding clone chimerism. (N, column 3) STSs from both YAC-insert ends can be amplified from chromosome 6. (Y, column 3) YACs from which at least one insert-end STS is not present on chromosome 6. The result of a search of the MIT/Whitehead database for hits by STSs from chromosomes other than chromosome 6 is reported in column 4, with the numbers referring to the respective chromosome. Outlined YACs constitute a minimal set of nine clones linking HLA-F near the distal end of the MHC to D6S276. The presence of an STS in a YAC is indicated by a + or by a solid square (▪), the latter marking a clone-end STS at the end of the YAC from which it was derived. The absence of an STS in a YAC is indicated by a −. D6S1016 gives rise to PCR products that fall into a large and a small size class, the presence of which are denoted by ▵ and ▿, respectively. With the exception of y906h11, all YACs have been tested for all STSs.
A minimum of nine YACs is required to form a continuous clone path that links the MHC class I region with D6S276. YAC y225b1 at the centromeric end of the clone contig contains D6S265 and the HLA-A andHLA-G genes and is part of a published YAC contig across the entire MHC (Abderrahim et al. 1994). An STS specific for theHLA-F gene is present in clones y903b9 and y800g3, proximal to the myelin/oligodendrocyte glycoprotein gene (MOG; Gardinier et al. 1992). The gene encoding the RET finger protein (RFP; Takahashi et al. 1988) is located farther telomeric near the right end of y903b3.
D6S306, D6S105, and D6S464, a group of well-characterized polymorphic markers, each with one allele in linkage disequilibrium with hemochromatosis (Raha-Chowdhury et al. 1995), are located within the left third of y950h11, approximately at the center of the map. D6S1260 (Raha-Chowdhury et al. 1995), one of the most telomeric markers reported to be tightly linked to hemochromatosis before the publication by Feder et al. (1996), and D6S1016, a tetranucleotide-repeat marker thought to mark the telomeric boundary of the region of linkage disequilibrium with hemochromatosis (Seese et al. 1996), are other noteworthy landmarks on this YAC.
The hemochromatosis gene HLA-H is located in a map segment shared by clones y899g1 and y974a2. D6S299, a marker that has been placed 4 cM telomeric of D6S276 on the meiotic linkage map (Dib et al. 1996), was found to be within y935a8 and y874b3, unseparable from D6S276 based on the STS content of YAC clones at the telomeric end of the contig map.
The depth of clone coverage is highly variable along the YAC contig, reaching a maximum of 10 clones at D6S258. In contrast, only one YAC, y731a5, was found to be positive for D6S2216 and D6S2215. This weak link has been reinforced with large insert bacterial clones (see below). Outside this map segment of single YAC clone coverage, all but seven adjacent pairs of STSs (indicated by triple bars in Fig. 1) are connected by at least two independent YAC clones.
The STS content data for YAC clones extending centromeric of sy669h7-R contain many inconsistencies, and an unusually high fraction of YACs from this region (7/19) is represented as noncontiguous blocks of STSs. As noncontiguity violates the very basis of STS content mapping, the exact order of STSs in this map segment is poorly established.
Assuming the marker order in Figure 1 is correct, there are two spots that are apparently difficult to clone in YACs: The DNA aroundHLA-F and MOG, which is missing in at least two independent clones (y960h11 and y906h11), and the segment between STSs sy414g1-L and sy729b12-L, which is likely to represent an actual gap in clone coverage.
All three YACs known to contain STSs flanking this gap (y854f5, y814d10, and y849f12) appear to have suffered an internal deletion. y854f5 represents a curious cloning artifact in that both insert ends originate from this region of chromosome 6 but most of the intervening sequence has been replaced by a DNA fragment carrying a string of eight STSs from chromosome 14 (S. Schneider, pers. comm.). y814d10 is missing only one STS. This STS, which was derived from the end of y900g10, is not single-copy and can be amplified from chromosomes 2, 6, and 12. Therefore, an alternative explanation for the absence of this marker in y814d10 is chimerism of y900g10.
Telomeric of sy669h7-R, only three YACs (y733e8, y912c11, and y947f6) are listed as noncontiguous blocks of STSs. The most interesting case is y947f6, which links STS WI-3111, located centromeric ofHLA-H, to WI-3878, D6S1621, and D6S1281 but is deleted for the immediate region around HLA-H including all of YAC y899g1, which is ∼900 kb in size.
Fifty-three percent (23/43) of the YACs have at least one end STS that cannot be PCR amplified from DNA of a rodent cell line containing human chromosome 6, or are hit by non-chromosome-6 STSs in the MIT/Whitehead STS content database (see Fig. 1). These YACs are likely to be chimeric. Their insert sizes are therefore unreliable for estimating actual physical distances based on STS content information but provide an upper limit for distance estimates.
RecA-assisted restriction endonuclease (RARE) cleavage assays (Ferrin and Camerini-Otero 1991) specific for the EcoRI sites at the insert termini of selected YACs were developed, and the extent of clone overlap among a minimal set of YACs was determined (Gnirke et al. 1994). According to this analysis (data not shown), the entire YAC contig represents ∼8 Mb of DNA, not counting the DNA missing in y960h11 and y814d10 (Feder et al. 1996).
Bacterial Clone–Contig Maps
To obtain cloned DNA in a form that is easier to manipulate than YAC clones and to confirm the YAC-based map, we assembled an overlapping set of bacterial clones from human genomic libraries. These clones provide coverage in large-insert plasmids of the DNA between the STSs sy814d10-R and sy899g1-R. The bacterial clone contig was built in three stages, using different approaches for different sections.
The DNA between sy814d10-R and D6S105 was covered for the most part by PCR-based genome walking in a total human P1 library starting with clones containing D6S105, D6S306, sy871g6-L, and sy753h12-L. A small gap in P1-clone coverage between D6S105 and D6S306 was bridged by a λ phage (L-106 in Fig. 2).
Bacterial clone–contig and STS map encompassing ∼3 Mb of DNA between sy814d10-R and sy899g1-R. One hundred ten bacterial clones and their approximate size (if known) are indicated in the two leftmost columns. P1, PAC, and BAC clones are denoted by the prefixes p, pc, and b, respectively, followed by their respective library address. L-106 is a λ-phage clone. One hundred fifty-three STSs are indicated at the top. STSs in boldface type have not been described previously. STSs marked with a solid circle (•) are included in the YAC-based STS-content map in Fig. 1. The STSs were ordered so as to minimize the number of clones with noncontiguous blocks of STSs. Groups of STSs not uniquely ordered based on the STS content of the bacterial clones are indicated by solid bars below the STS names. Clones are depicted as shaded horizontal bars. FISH-mapped clones mentioned in the main text are outlined. The presence of an STS in a clone is indicated by a + except for a clone-end STS at the end of the clone from which it was derived (▪), and for D6S1016, which gives rise to a large (▵) and a small (▿) size class of PCR products. A − denotes the absence of the an STS in a clone. Empty cells have not been determined but are expected to be negative. The portion of the matrix within the rectangle at the upper leftcorner contains only a minimal number of clones, the STS content of which has been determined in part by “electronic PCR,” that is, by searching (in some cases, incomplete) clone-specific sequence libraries for STS or primer sequences. The shaded map segment indicates a consecutive block of eight STSs that can be amplified from chromosome 5 and 6. The approximate extent of the region that is duplicated near the chromosome-6 centromere is indicated (see main text).
To cover the region between D6S105 and sy950h11-R, a y950h11-derived cosmid library was constructed that served as the starting material for the generation of 14 hybridization probes and STSs, 6 of which contained (CA)n repeats. These STSs were used to screen P1, bacterial artificial chromosome (BAC), and P1-derived artificial chromosome (PAC) libraries. Additional P1 clones from this genome region were identified by hybridization with inter-AluPCR products from YAC y950h11. Initially, five short contigs were obtained that were expanded by bidirectional walking using STSs and hybridization probes developed from the end of clone inserts. Remaining gaps were closed by generating an additional 17 random STSs from gel-purified y950h11 DNA followed by STS content mapping and screening with selected STSs at the ends of contigs.
Bacterial clone coverage across the genomic region represented by YAC y899g1 was obtained by screening BAC and PAC libraries for 26 random y899g1-derived STSs and four YAC-end STSs (sy899g1-L, sy950h11-R, sy974a2-L, and sy899g1-R). The resulting clone collection was supplemented with P1 and BAC clones found by hybridization to a whole-YAC probe generated by inter-Alu PCR. A comprehensive STS content matrix of the 48 clones identified by both screening methods resulted in a single contig of 41 clones spanning at least 900 kb, the size of y899g1. Seven clones that were positive by hybridization with the inter-Alu–PCR probe were not hit by any STS and therefore not incorporated in the final map. For this section of the bacterial clone contig, STSs generated from the ends of clone inserts were only used for map confirmation and refinement rather than contig expansion.
An STS content map comprising 110 bacterial clones (38 BACs, 45 PACs, 26 P1 clones, and one λ clone) and 153 STSs is shown in Figure 2. This map represents genomic DNA between the right end of YAC y814d10 and the right end of YAC y899g1, which are ∼3 Mb apart (Feder et al. 1996). The centromeric end of the bacterial clone contig provides independent confirmation of the region of single-YAC clone coverage (see above) with P1 clone p1079d10 linking STSs sy814d10-R through D6S2216, and P1 clone p950a12 containing D6S2216, D6S2215, and sy669h7-R. In the map representation shown in Figure 2, four bacterial clones (b52n11, b109e23, pc81a8, and b58o10) have a noncontiguous complement of STSs, indicating either clone deletion artifacts or polymorphisms present in the source DNAs from which the clone libraries were constructed. The HLA-H gene is located in the region of overlap between clones b132a2 and pc222k22 within the rightmost quarter of the map.
A Multicopy Genome Segment
The most striking feature of the bacterial clone-based STS content map is a tall stack of clones with essentially identical STS content near the center of the contig (see map segment flanked by sy950h11-22 and sy899g1-L in Fig. 2). To test the hypothesis that this phenomenon is attributable to a duplication elsewhere in the genome, we tested the PCR assays on a panel of monochromosomal hybrid cell lines. Surprisingly, all STSs in the region of extreme depth of clone coverage are specific for chromosome 6. However, a contiguous block of eight STSs directly adjacent to the stack of clones amplified from chromosome 5 as well.
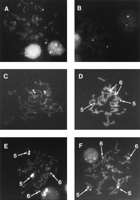
To determine the chromosomal origin of clones within and adjacent to the clone stack, we performed fluorescence in situ hybridization (FISH) on metaphase and interphase nuclei using Cy3-labeled large insert plasmid DNA as probes. As a control, two clones (b28a18 and p1065b2) flanking this segment were used. Both control probes hybridized to a single target region on chromosome 6p (Fig. 3A,B), the identity of which was verified by dual-color hybridization using Cy3-labeled plasmid and a FluorX-labeled chromosome 6-specific α-satellite probe (data not shown). In contrast, p20p19 gave rise to two distinct hybridization signals on chromosome 6 (Fig. 3C). On cohybridization with the α-satellite probe, the second hybridization signal of the p20p19 probe could not be resolved from the α-satellite block on metaphase chromosomes, and both probes appeared to hybridize immediately adjacent to each other in interphase nuclei as well (data not shown). This finding suggests that the second hybridization target lies close to the chromosome-6 centromere. b367j20, a clone that extrudes from the stack into the region duplicated on chromosome 5, gave a weak signal on chromosome 5 in addition to the double signal on chromosome 6 (Fig. 3D). Additional multiple spots on chromosome 5 were visible after probing with b19e12, which lies in the center of the promiscuous DNA segment (Fig. 3E). Another clone, pc23i1 (Fig. 3F), hybridized to both chromosome 5 and 6, but did not give rise to a clearly visible hybridization signal near the chromosome-6 centromere.
FISH analysis of probes representing six bacterial clones. Chromosome spreads were hybridized with Cy3-labeled plasmid DNA from clones b28a18 (A), p1065b2 (B), p20p19 (C), b367j20 (D), b19e12 (E) and pc23i1 (F). Chromosomes 6 and 5 were discriminated in parallel dual-color hybridization experiments using Cy3-labeled large-insert plasmid DNA and FluorX-labeled chromosome-6 or chromosome-1, 5 and 19-specific alphoid-DNA probes (data not shown). The hybridization signal inA, B, and C is on chromosome 6.
Although the region that is duplicated on chromosome 5 can be recognized easily as a block of eight STSs that can be amplified from a rodent cell line harboring human chromosome 5, it is not straightforward to define the boundaries of the zone that is duplicated elsewhere on chromosome 6. The closest flanking clones that were tested by FISH and did not hybridize to two distinct regions on chromosome 6 were pc23i1 and b157e10. The latter is a chimeric BAC whose Sp6 end is from chromosome 15 but whose chromosome-6 portion hybridized to a single locus on chromosome 6p (data not shown).
We also employed radiation hybrid (RH) mapping to determine the chromosomal and subchromosomal origin of STSs from this region (see Table 1). Three STSs immediately centromeric to the segment duplicated on chromosome 5 are RH-linked to markers D6S1852 or D6S1558, an STS that has been placed next to WI-3111 on the MIT/Whitehead YAC-based STS content map (Hudson et al. 1995). Both linked STSs fall within RH map bin 19. Similarly, three STSs immediately telomeric to the clone stack are RH linked to markers in bin 19 or the neighboring bin 18. In contrast, most intervening STSs display RH linkage to markers in bins 58 or 59, located close to the chromosome-6 centromere. The only exception is sp202l10–Sp6, which hits D6S1986 in bin 73 and is the only STS displaying linkage to chromosome-5 markers at a lod score of >3. All intervening STSs hit at least 34 RH samples, more than twice the average number of hits reported for this RH panel (D.R. Cox, pers. comm.). By using >30 hits as an arbitrary criterion for an STS that is present in more than one copy, the multicopy region is confined to the interval between STSs sy950h11-18 and sy899g1-L.
Characterization of Selected STSs in and Around the Multicopy Genome Segment by RH Mapping
An interesting case is the tetranucleotide-repeat marker D6S1016 that gives rise to two distinct PCR products (∼300 and 220 bp in size, respectively) on an agarose gel. The marker that is RH linked most tightly to the smaller D6S1016 product is D6S2014 near the chromosome-6 centromere, whereas the larger product is linked to D6S1558 in this region of chromosome 6. Therefore, the two size classes represent different loci rather than alleles. All three bacterial clones harboring D6S1016 and at least one unique STS from this region of the chromosome (b297d15, p20p19, pc17k19) give rise to the larger size D6S1016 PCR product. This rule holds for YACs y950h11, y912c11, and y905g1 as well (see Fig. 1). YAC y836c6, on the other hand, is likely to have originated from the region near the chromosome-6 centromere; it contains the smaller D6S1016 STS, and both y836c6-end STSs can be amplified from chromosome 6 but not from any other clone in our collection.
The presence of this multicopy genome segment explains some inconsistencies we noted during the assembly of the bacterial clone contig. For example, a short contig of five P1 clones seemed to overlap perfectly with several other clones. However, three consecutive STSs at the other end of that contig could not be accommodated in the final STS content map without formation of a branched map. These three STSs were neither present in YAC y950h11 nor y912c11, whereas otherwise both YACs contained all STSs from this region (data not shown). Another inconsistency in this segment of the STS content map is that not all clone-end STSs can be placed at the very end of the clone from which they have been developed (see Sp6 ends of b55c20 and b109e23 in the map representation shown in Fig. 2).
No bacterial clone path can be constructed that does not involve at least two clones that are contained fully within the duplicated region. BAC b19e12, for example, is an obligatory member of the clone contig but did not necessarily originate from chromosome 6. Furthermore, all possible tiling paths include either b55c20 or b109e23, two clones that harbor the small D6S1016 STS. Therefore, it remains uncertain whether all DNA from this region on 6p21.3–p22 has been accounted for in bacterial clones.
DISCUSSION
We have constructed a YAC clone and a bacterial clone-based STS content map that served as framework physical maps for the positional cloning of the HLA-H gene, a candidate gene for hereditary hemochromatosis (Feder et al. 1996). The YAC contig represents some 8 Mb of human chromosome 6 between the class I region at the telomeric end of the MHC in 6p21.3 and D6S276, located in 6p22 (Bray-Ward et al. 1996). Approximately 3 Mb of this genome segment was recovered as a single large insert bacterial clone contig. Together, the YAC and bacterial clone-based STS content maps comprise 197 different STSs, 47 of which are polymorphic microsatellite markers, and 140 of which have not been described previously. The two maps have 42 STSs in common. These shared STSs form a contiguous block in the YAC-based map and allow the two STS content maps to be aligned along the entire length of the bacterial clone contig.
The centromeric end of the YAC contig overlaps with a published YAC contig across the MHC (Abderrahim et al. 1994). Our results are consistent with two YAC contigs published recently, one constructed byBurt et al. (1996), which covers the region from the telomeric end of the MHC up to and including YAC y950h11, and one by Malaspina et al. (1996), which extends from D6S306 to the telomeric end of our map and beyond. The contig map constructed by Burt et al. (1996) lists 16 YACs from three different libraries and suggests a similar tiling path of CEPH YACs, except for the region that appears to be under-represented in the CEPH library (D6S2216 and D6S2215 in our map), which has been bridged by an ICI YAC (Anand et al. 1990) instead of the CEPH clone y731a5. The YAC contig map by Malaspina et al. (1996) is deeper in clone coverage than the one presented here; the overlap relationships among the clones that their and our map have in common are essentially the same.
The region of minimal clone coverage in the CEPH YAC library marks the gap between contigs WC6.0 and WC6.1 in the MIT/Whitehead map (T. Hudson, L. Stein, S. Gerety, J. Ma, A. Castle, J. Silva, D. Slonim, R. Baptista, L. Kruglyak, S. Xu et al. An STS-based map of the human genome. MIT/Whitehead database release 11, October 1996), which otherwise deviates from our map and that by Malaspina et al. (1996) in several respects. The most obvious discrepancy affects the region in the immediate vicinity of the HLA-H gene. Telomeric of y905g1, our results indicate an obligatory tiling path through clones y899g1 and y974a2, whereas neither of these two YACs has been incorporated into the WC6.0 contig. Instead, YACs y950h11, y912c11, and y905g1 have been connected via the internally deleted YAC y947f6 to a group of more telomeric clones including y792g12 and y901a10, thereby skipping the genome segment harboring the HLA-H gene. Because of the DNA missing in y947f6, physical distances in this genomic region have been underestimated (Raha Chowdhury et al. 1996). Similarly, in the CEPH YAC contig map, both YACs y899g1 and y974a2 are lacking in the bins at chromosome-6 map positions 0.42 and 0.46 (Chumakov et al. 1995). The map by Malaspina et al. (1996) lists several clones that may substitute for y899g1 and y974a2, none of which have been incorporated in the CEPH or MIT/Whitehead contig map. As a result, the HLA-H gene was not represented in any public YAC contig, probably impeding the search for the hemochromatosis gene in laboratories solely relying on public clone–contig maps.
A peculiar feature of this genomic region is the presence of a segment that is duplicated elsewhere on chromosome 6 and, in part, on chromosome 5 as well. Originally suspected by the unusual depth of bacterial clone coverage and by inconsistencies noted during the contig assembly process, the particularly insidious duplication within the same chromosome was revealed by FISH analysis of bacterial clones and RH mapping of selected STSs. Both of the copies near the chromosome-6 centromere and on chromosome 5 must be of high enough sequence similarity to this segment on chromosome 6p21.3–p22 to allow cross-amplification of several PCR assays and cross-hybridization at the stringency of a FISH experiment.
Because the multicopy DNA segment is more than one bacterial clone-length in size, it represents a formidable obstacle for physical mapping with bacterial clones. We were unable to construct a single bacterial clone path, where all members are bona fide representatives of this region of chromosome 6. Choosing a minimal set of bacterial clones for further studies of this DNA segment will require the identification of region-specific variants of STS sequences or restriction fingerprinting of the bacterial clones themselves. The larger YACs are clearly superior for mapping across a problem zone like this, and direct subcloning of YAC DNA may prove more practical than assembling a contig of bacterial clones from a total genomic library.
Because it contains multicopy DNA, YAC y950h11 has been described as chimeric based on both FISH (Bray-Ward et al. 1996) and inter-Alu probe hybridization evidence (Chumakov et al. 1995). Yet, both insert ends originate from this region of chromosome 6p, and, by other criteria including STS content and the distance in uncloned DNA between the EcoRI sites at the YAC insert termini as determined by RARE cleavage (A. Gnirke, unpubl.), y950h11 appears to carry a faithful copy of the corresponding genomic DNA.
In addition to posing a challenge for clone-based physical mapping, the multicopy DNA segment that contains the polymorphic D6S1016 marker may have confounded genetic analyses as well. It was at this marker where the linkage disequilibrium with hemochromatosis was reported to drop, therefore suggesting a telomeric boundary of the search area for the hemochromatosis gene (Seese et al. 1996).
The clone–contig and STS maps reported here will facilitate the general characterization of this region of the human genome and may be valuable in future searches for genes involved in human disease. Currently, the list of potential susceptibility loci that may fall in or near this region of chromosome 6 include loci for dyslexia (Cardon et al. 1994; Grigorenko et al. 1997), schizophrenia (Schwab et al. 1995), asthma (Daniels et al. 1996), ovarian carcinoma (Foulkes et al. 1993), and numerous diseases that display association with polymorphic markers within the MHC.
METHODS
YACs
YAC library addresses were identified by querying the MIT/Whitehead (Hudson et al. 1995) and CEPH-Généthon databases (Chumakov et al. 1995), and by PCR-based screening of plates 613–984 of the CEPH YAC library (Dausset et al. 1992), which was purchased from Research Genetics (Huntsville, AL). At least four different clone isolates were analyzed from each positive well, and the largest clone containing the STS of interest was studied further. YAC clones are designated by their address in the CEPH library preceded by the prefix y. DNA from YAC clones in agarose blocks was prepared using a modification of the method of Southern et al. (1987). YAC sizes were determined by pulsed-field gel electrophoresis and hybridization to vector-specific probes using λ concatemers and Hansenula wingei chromosomes (Bio-Rad, Hercules, CA) as size standards. For PCR, an aliquot of the agarose block was diluted with two volumes of H2O, melted, and liquified by digestion with β-agarase I (New England Biolabs, Beverly, MA). YAC insert ends were isolated by a modification (Green 1993) of the vectorette PCR method (Riley et al. 1990). PCR products were gel purified and sequenced using standard fluorescent dye terminator chemistry (Perkin Elmer, Norwalk, CT). Pulsed-field gel-purified YAC DNA was isolated as described (Gnirke et al. 1993) and cleaned up by phenol extraction and ethanol precipitation. Random sequences from purified YAC DNA for STS development were obtained by constructing complete EcoRI,SacI, and EcoRI–SacI digest libraries in pBluescript SK (Stratagene, La Jolla, CA) or RsaI andHaeIII digest libraries in pSP72 (Promega, Madison, WI), and sequencing the insert ends of randomly picked plasmid clones. To eliminate redundancies and contaminating yeast sequences, the sequences were compared both to each other and to the nonredundant sequence division of GenBank by BLAST (Altschul et al. 1990). Whole-YAC hybridization probes were made by long-range inter-Alu PCR (D.L. Nelson, pers. comm.) of DNA from YAC-containing yeast strains and radio-labeled by standard procedures (Feinberg and Vogelstein 1983).
Large-Insert Bacterial Clones
P1 clones were identified by hybridization or PCR-based library screening (Genome Systems, St. Louis, MO). PACs containing STSs of interest were identified by PCR screening of DNA pools from a total human PAC library (Genome Systems). BAC clones were from plates 1–384 of a total human BAC library purchased from Research Genetics. Numbers of BAC library plates containing positive clones were determined by PCR screening of DNA plate pools (Research Genetics). Complete BAC library addresses were determined by testing row-and-column pools of liquid cultures grown in a 384-well plate by PCR. Bacterial clones are designated by the coordinates of positive wells in the respective Genome Systems or Research Genetics clone collection preceded by the prefixes p, pc, or b for P1s, PACs, or BACs, respectively. The STS content of bacterial clones was determined by diluting bacterial cultures 1:100 in the PCR reaction mixture. Small-scale large-insert plasmid preparations were performed using an alkaline lysis protocol (B. Birren, pers. comm.). To obtain end sequences of bacterial clone inserts, P1, PAC, or BAC DNA was first digested with a restriction enzyme that does not cleave within the vector (NdeI or SacI for P1s, and PACs; NsiI,BbrPI or NheI for BACs), followed by recircularizing, and direct sequencing of the resulting plasmids of reduced complexity using vector-specific sequencing primers. Screening of a library of total human DNA in Lambda DASH II (Stratagene) by hybridization was performed by using standard procedures.
STSs and PCR Conditions
Repetitive sequences detected by comparison with a database containing human repetitive sequence elements were masked before choosing PCR primers by OSP (Hillier and Green 1991). YAC-derived STSs are designated by the prefix s followed by the name of the YAC it was derived from, and either a consecutive number (random STSs) or L or R [clone-end STSs adjacent to the left (centric) or right (acentric) YAC-vector arm, respectively]. Bacterial clone-derived STSs are designated by the prefix s followed by the name of the bacterial clone it was derived from and either a consecutive number (random STSs) or T7 or Sp6 (clone-end STSs). sHHp61 and sHHp89 are STSs that contain a single-base-substitution polymorphism (see GDB database entries for D6S2310 and D6S2316, respectively). STSs were amplified in either PTC 200/225 (MJ Research, Watertown, MA) or PE9600 thermocyclers (Perkin Elmer), using one standard reaction buffer (GeneAmp with 1.5 mm MgCl2; Perkin Elmer) but variable, empirically determined annealing temperatures. Standard cycling conditions for the PTC 200/225 were a 2-min initial denaturation step at 92°C followed by 35 cycles of 20 sec at 92°C, 45 sec at the annealing temperature and 1 min at 72°C, and a final one-time 2-min incubation at 72°C. Standard cycling conditions on the PE9600 were 1 min at 94°C followed by 35 cycles of 20 sec at 94°C, 20 sec at the annealing temperature and 30 sec at 72°C, and a final 2-min incubation at 72°C. Annealing temperatures in the PTC 200/225 thermocyclers were 50°C, 55°C, or 60°C. For the PE9600 instruments, the annealing temperatures were raised by 5°C. Both thermocycling regimes were used interchangeably. The chromosomal origin of STSs was determined using the National Institute of General Medical Sciences (NIGMS) somatic-cell-hybrid-mapping panel 2, version 2 (Coriell Institute, Camden, NJ). Primer sequences and size ranges of STSs with a D6S, WI, or a tetranucleotide P designation are available from the GDB, the MIT/Whitehead, and the CHLC databases, respectively. Primer sequences for the amplification of HLA-A, HLA-G, andHLA-F were from Abderrahim et al. (1994). MOG, BT, and RFP primers have been described by Amadou et al. (1995). Primer pairs specific for HLA-H were as described (Feder et al. 1996). Primer sequences, PCR product sizes, and annealing temperatures for PCR assays that amplify more than one locus in the human genome as well as for two STSs from the ends of y836c6 are given in Table 2. All other STSs have been entered into the STS division of GenBank and the GDB database under the accession numbers indicated in Figures 1 and 2.
PCR Primers, Product Sizes, and Annealing Temperatures for Multicopy Amplicons and STSs Outside This Region of Chromosome 6p21.3–p22
FISH
Human metaphase chromosomes were prepared using standard procedures (Yunis 1976). Large-insert plasmid DNAs were purified from 500-ml cultures using a commercial iron-exchange resin (tip 500, Qiagen, Chatsworth, CA), and 1 μg was directly labeled with Cy3–dCTP by nick translation (FluoroLink Cy3 nick translation kit; Amersham, Arlington Heights, IL). One hundred fifty nanograms of labeled probe was prehybridized in a 20 μl volume in 50% formamide, 10% dextran sulfate, and 2× SSC (pH 7.0) with 2 μg C0t-1 DNA (GIBCO-BRL, Gaithersburg, MD) at 37°C for 30 min and hybridized to metaphase chromosomes on microscope slides under a 24 × 50-mm coverslip overnight at 37°C in a humid chamber. FluorX-labeled α-satellite probes (Amersham) specific for chromosome 6 or chromosomes 1, 5, and 19 were cohybridized with Cy3-labeled plasmid probes to confirm chromosomal localization. Slides were washed three times, 5 min each, at 42°C in 50% formamide, 2× SSC (pH 7.0) followed by three 5-min washes at 42°C in 2× SSC (pH 7.0), rinsed in phosphate-buffered detergent (Pinkel et al. 1986), counterstained with DAPI, mounted in antifade solution, and inspected on a Zeiss Axioskop fluorescence microscope. Separate DAPI, FluorX, and Cy3 pictures were taken with Ektachrome 320T slide film (Kodak, Rochester, NY).
RH Mapping
Characterization of selected STSs by RH mapping was performed using the Stanford G3 RH mapping panel, which was obtained from Research Genetics. The resulting STS content data were submitted to the World Wide Web server of the Stanford Human Genome Center for two-point maximum-likelihood analysis.
Acknowledgments
We thank Denis Le Paslier for replacing YAC clones that failed to grow from our copy of the CEPH YAC library, David Nelson for an unpublished protocol for long-range inter-Alu PCR, Bruce Birren for a surprisingly simple large-insert plasmid miniprep protocol, John Feder for contributions to the early stages of this project, the Mercator genotyping group for (CA)n-repeat markers, the Mercator sequencing group for sequencing of PCR products and plasmid subclones, Gerard Bouffard and Melissa Lukes for help with STS submission, and Mike Ellis, Sandy Schneider, Winston Thomas, and Zenta Tsuchihashi for helpful discussions and critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.