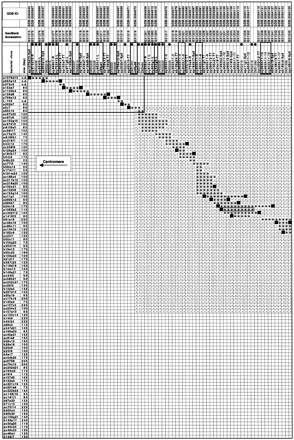
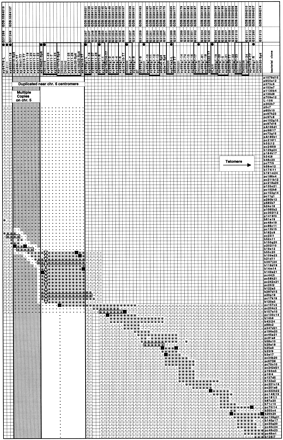
Bacterial clone–contig and STS map encompassing ∼3 Mb of DNA between sy814d10-R and sy899g1-R. One hundred ten bacterial clones and their approximate size (if known) are indicated in the two leftmost columns. P1, PAC, and BAC clones are denoted by the prefixes p, pc, and b, respectively, followed by their respective library address. L-106 is a λ-phage clone. One hundred fifty-three STSs are indicated at the top. STSs in boldface type have not been described previously. STSs marked with a solid circle (•) are included in the YAC-based STS-content map in Fig. 1. The STSs were ordered so as to minimize the number of clones with noncontiguous blocks of STSs. Groups of STSs not uniquely ordered based on the STS content of the bacterial clones are indicated by solid bars below the STS names. Clones are depicted as shaded horizontal bars. FISH-mapped clones mentioned in the main text are outlined. The presence of an STS in a clone is indicated by a + except for a clone-end STS at the end of the clone from which it was derived (▪), and for D6S1016, which gives rise to a large (▵) and a small (▿) size class of PCR products. A − denotes the absence of the an STS in a clone. Empty cells have not been determined but are expected to be negative. The portion of the matrix within the rectangle at the upper leftcorner contains only a minimal number of clones, the STS content of which has been determined in part by “electronic PCR,” that is, by searching (in some cases, incomplete) clone-specific sequence libraries for STS or primer sequences. The shaded map segment indicates a consecutive block of eight STSs that can be amplified from chromosome 5 and 6. The approximate extent of the region that is duplicated near the chromosome-6 centromere is indicated (see main text).











