The Comparative Genomic Structure and Sequence of the Surfeit Gene Homologs in the Puffer Fish Fugu rubripes and their Association with CpG-Rich Islands
Abstract
The puffer fish Fugu rubripes (Fugu) has a compact genome approximately one-seventh the size of man, mainly owing to small intron size and the presence of few dispersed repetitive DNA elements, which greatly facilitates the study of its genes at the genomic level. It has been shown previously that, whereas the Surfeit genes are tightly clustered at a single locus in mammals and birds, the genes are found at three separate loci in the Fugu genome. Here, Fugu gene homologs of all six Surfeit genes (Surf-1 to Surf-6) have been cloned and sequenced, and their gene structure has been compared with that of their mammalian and avian homologs. The predicted protein products of each gene are well conserved between vertebrate species, and in most cases their gene structures are identical to their mammalian and avian homologs except for the Fugu Surf-6 gene, which was found to lack an intron present in the mouse gene. In addition, we have identified conserved regulatory elements at the 5′ and 3′ ends of theSurf-3/rpL7a gene by comparison with the mammalian and chickenSurf-3/rpL7a gene homologs, including the presence of a polypyrimidine tract at the extreme 5′ end of this ribosomal protein gene. The Fugu Surfeit gene homologs appear to be associated with CpG-rich islands, like the Surfeit genes in higher vertebrates, but these Fugu CpG islands are similar to the nonclassical islands characteristic of other fish species. Our observations support the use of the Fugu genome to study vertebrate gene structure, to predict the structure of mammalian genes, and to identify vertebrate regulatory elements.
[The sequence data described in this paper have been submitted to the data library under accession nos. Y15170 (Surf-2, Surf-4), Y15171(Surf-3, Surf-1, Surf-6), and Y15172(Surf-5.)]
The genome of the Japanese puffer fish,Fugu rubripes (Fugu), is ∼7.5 times smaller than the human genome (400 Mb compared with 3000 Mb) mainly owing to the presence of fewer dispersed repetitive DNA elements and smaller introns (Brenner et al. 1993). Despite being compact, the Fugu genome is thought to possess a similar gene repertoire to other vertebrates and has therefore been proposed as a model genome for studying vertebrate gene structure. An increasing number of Fugu gene homologs have been identified that are highly homologous to their mammalian counterparts at the amino acid level and also show a conserved gene structure (e.g., intron/exon boundaries) (Baxendale et al. 1995; Elgar et al. 1995; Mason et al. 1995; Venkatesh and Brenner 1995; Maheshwar et al. 1996; Venkatesh et al. 1996). Furthermore, sequence comparisons between the noncoding DNA of Fugu genes and their higher vertebrate homologs have revealed conserved elements thought to be required for gene regulation or associated with other functions such as intron-encoded small nucleolar RNAs (Aparicio et al. 1995; Marshall et al. 1994; Cecconi et al. 1996; Crosio et al. 1996).
The mouse Surfeit locus contains at least six sequence-unrelated genes (Surf-1 to Surf-6) and encompasses ∼45 kb of genomic DNA (Huxley and Fried 1990b). The six Surfeit genes have been classified as housekeeping genes, being expressed in all tissue types tested and not containing a TATA box in their promoter region. The mouse Surfeit locus contains four CpG-rich islands that are associated with the 5′ ends of the six Surfeit genes (Fig.1A). The relatively high gene density within the Surfeit locus (an average of one gene every 7.5 kb) compared with the mouse genome as a whole, the alternation of transcription of five of the genes with respect to their neighbors, the presence of one confirmed bidirectional promoter (that of the Surf-1 andSurf-2 genes), and the overlap of two of the Surfeit gene transcripts have led to the suggestion that the unusual gene organization may have regulatory and/or functional significance (Huxley and Fried 1990b; Gaston and Fried 1994; Lennard et al. 1994). At present, only the function of the Surf-3 gene, which encodes the ribosomal protein L7a (Surf-3/rpL7a gene), is known (Giallongo et al. 1989), although it is additionally known that theSurf-4 gene encodes an integral membrane protein of the endoplasmic reticulum (Reeves and Fried 1995), that the Surf-6gene encodes a novel nucleolar protein (Magoulas and Fried 1996), and that an Saccharomyces cerevisiae gene homologous to the mammalian Surf-1 gene encodes a mitochondrial protein required for respiration (Mashkevich et al. 1997).
Genomic organization of the mouse Surfeit locus and comparison with the organization of the Fugu Surfeit genes at three separate loci. (A) Organization of the mouse Surfeit locus. The relative orientation of the genes and their intergenic distances are shown (adapted from Garson et al. 1995). The continuous line represents genomic DNA. The direction of transcription of each gene (Surf-1 to Surf-6) is indicated by arrows, and the 5′ end of each gene can be seen to be associated with a CpG island (▪). In human the ASS gene is located ∼2–4 Mb from the Surfeit locus 3′ to the Surf-6 gene. The human EST00098 has been mapped to chromosome 9 but is not found within 50 kb either side of Surfeit locus as determined by PCR. These features have not been determined in mouse. (B) Cosmid contig construction demonstrates that the Fugu Surfeit gene homologs are located at three separate loci (modified from Gilley et al. 1997). Cosmid contigs were constructed around each of the three Fugu genomic loci; (i) containing the Fugu Surf-3/rpL7a, Surf-1, and Surf-6 gene homologs, (ii) containing the FuguSurf-2, Surf-4, and ASS gene homologs and sequences homologous to human EST00098, and (iii) containing the Fugu Surf-5 gene homolog only. In each case, Fugu genomic DNA is represented as a thick horizontal line, and, above, the direction of transcription and position of each of gene is expanded for clarity. Intergenic distances between Fugu Surfeit genes are shown. Cosmid contigs are highlighted below the genomic DNA with each cosmid clone shown as a thin horizontal line. Each cosmid is labeled with its original Fugu cosmid library clone number. Cosmids in each contig were isolated and arranged relative to one another using restriction and hybridization analyses. None of the cosmids overlap with cosmids from either of the other two contigs. The approximate distance that each contig stretches either side of the FuguSurfeit gene loci is shown in kilobases below the genomic DNA. Each contig shows only informative cosmids. Additional cosmids in contigi are 006I18, 041C23, 059G12, 070P11, 111D07, 111M11, 111N11, 117M14, 194B16, and 194C20, and additional cosmids in contigii are 007P02, 028B15, 044B19, 139G10, 186G08, and 196C01. No additional cosmids could be identified for locus iii, owing to the probable presence of numerous repetitive elements.
The unique spatial arrangement of at least five of the Surfeit genes (Surf-1 to Surf-5) has been shown to be conserved between mouse, human, and chicken (Colombo et al. 1992; Yon et al. 1993), whereas at least five of the Surfeit genes are not linked in the two invertebrate species tested, Drosophila melanogaster andCaenorhabditis elegans (Armes and Fried 1995, 1996). Tetraodontoid fish are estimated to have diverged from the mammalian and avian lineages ∼430 million years ago, which is conveniently positioned midway between the divergence points of the avian (300 million years) and invertebrate (600 million years) lineages from the mammalian lineage. Fugu should therefore be informative in the evolutionary analysis of the Surfeit locus and its genes.
We have reported previously that Fugu homologs of all six Surfeit genes have been isolated, that they are represented only once in the Fugu genome, and that their genomic organization is largely different from that found in higher vertebrates being located at three separate loci in the Fugu genome (Fig. 1) (Gilley et al. 1997). Even when Fugu Surfeit genes are found together, their gene order is largely different from that found in mammals and birds. In this paper we have analyzed the conservation of gene structure between the six Fugu Surfeit gene homologs and their mammalian counterparts. We find that the structures of the six Surfeit genes are largely conserved between Fugu and higher vertebrates and that the predicted products of the six Fugugene homologs are highly homologous to the corresponding mouse proteins. In addition, we demonstrate that Fugu Surfeit gene homologs are associated with CpG-rich islands that we find are similar in composition to other characterized fish CpG islands (Cross et al. 1991). Otherwise, with the exception of the Surf-3/rpL7a gene, we can recognize little conservation of promoter elements between theFugu and mammalian Surfeit genes.
RESULTS
Isolation of F. rubripes Surfeit Gene Homologs
A gridded Fugu cosmid genomic library sufficiently complex to cover eight genomes was screened with cDNA inserts of the six mouse and/or human Surfeit genes (Surf-1 toSurf-6) (see Materials and Methods). Positively hybridizing cosmids were identified initially with mouse or humanSurf-3/rpL7a, Surf-4, and Surf-5 cDNA probes. Cosmids 036L10, 186H17, and 194D10 were identified with a Surf-3/rpL7aprobe, cosmids 007P02, 028B15, 044B19, 084H09, 139G11, 186G08, and 196C01 with a Surf-4 probe, and cosmids 028I13 and 177E10 with a Surf-5 probe. Cosmids 186H17, 139G11, and 177E10 were studied further by restriction enzyme and Southern blot analyses that subsequently revealed that sequences homologous toSurf-2 resided on the cosmids containing Surf-4homologous sequences and that sequences homologous to Surf-1and Surf-6 resided on cosmids containing Surf-3/rpL7ahomologous sequences. The extent and relative location of each of the six Fugu Surfeit gene homologs was determined by Southern blot analysis of cosmid restriction digests and sequence analysis of subcloned cosmid DNA (Fig. 1) (Gilley et al. 1997).
Conservation of the Intron/Exon Organization Between theFugu and Mouse Surfeit Gene Homologs
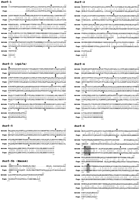
The putative intron/exon organization of each of the sixFugu Surfeit gene homologs within their coding regions was deduced from the structure of the corresponding mouse Surfeit genes. The position of the last three introns of the Fugu Surf-1 gene and the position of the last four introns of the Fugu Surf-3/rpL7a gene have been additionally confirmed following the isolation of three Fugu Surf-1 cDNA clones and 14 Fugu Surf-3/rpL7a cDNA clones from a directionally cloned [poly(A)-selected] Fugu 5′-stretch plus cDNA library (Clontech). No other informative Fugu Surfeit gene cDNA clones have been isolated. The structures of the Surf-2, Surf-3/rpL7a, and Surf-4 Fugu gene homologs, within their coding regions, are identical to the structures of the corresponding mouse genes with all intron/exon boundaries predicted to be conserved (Fig. 2). The Fugu Surf-1gene homolog also appears to share an identical structure to its mammalian counterparts from a position within the third exon to its termination codon. Before this position, no homology to the mammalian Surf-1 proteins is evident, and we have therefore not been able to define the extreme 5′ end of the Fugu Surf-1 gene homolog by comparison. The intron/exon organization of the Fugu Surf-6gene homolog differs from that of the mouse Surf-6 gene in that it possesses only three introns within the coding region of the gene, whereas the mouse gene possesses four (Magoulas and Fried 1996) (Fig.2). Two of the three Fugu Surf-6 introns are found in identical positions to the first and third introns of the mouse gene. The second intron in Fugu is predicted to be in a similar, but not identical, position to the mouse intron 2, and it should be noted that this region is very poorly conserved between mouse andFugu making it very difficult to predict the exact position of this intron/exon junction (Fig. 2). The fourth intron found in mouse is absent in Fugu (Fig. 2). Finally, the intron/exon organization of the Fugu Surf-5 gene homolog is confused by the fact that the mouse Surf-5 gene specifes two proteins (Surf-5 and Surf-5b) as a result of differential splicing (Garson et al. 1995,1996). The ubiquitous mouse Surf-5 3.5-kb mRNA that specifies an 140 amino-acid-protein and the mouse 1.5-kb Surf-5b mRNA that specifies the tissue-specific 200-amino-acid protein share three exons whose lengths and positions are conserved in the Fugu Surf-5 gene homolog (Fig. 2). The mouse Surf-5b mRNA contains a fourth exon that is derived from the 3′-untranslated region of the mouse Surf-5 mRNA. This exon encodes an additional 63 amino acids not found in the ubiquitous mouse Surf-5 protein. These additional 63 amino acids are less well conserved between mouse and human than the 137 amino acids that are common to both the Surf-5 and Surf-5b proteins (Garson et al. 1996). A search of the sequence downstream of the Fugu Surf-5 coding region reveals a small open reading frame specifying 68 amino acids that includes a stretch of 8 amino acids that are identical to a stretch of 8 amino acids in the additional 63 amino acids specific to the Surf-5b protein (Fig. 2). In addition, the donor and acceptor splice sites for the intron predicted for this putative additional exon are in conserved positions when compared with mouse. Therefore, the Fugu Surf-5gene homolog may also specify a Surf-5b protein by differential splicing.
Amino acid homology between the mouse and predicted FuguSurfeit gene products. The GAP program from the GCG software suite was used to generate amino acid sequence alignments comparing each of the six mouse Surfeit proteins with the predicted protein products of the six Fugu Surfeit gene homologs that have been deduced from the predicted structure of each Fugu gene. Vertical lines between the sequences indicate identity, double dots indicate conservation of similar amino acids, and single dots indicate changes found to occur frequently between homologs. Numeration of both proteins is given from their putative initiator methionines except for the putativeFugu Surf-1 protein whose amino terminus has not been elucidated and the Surf-5b protein for which alignment begins at amino acid 131 of both Fugu and mouse proteins. Vertical arrows above the mouse sequences and below the Fugu sequences indicate the relative positions of intron/exon boundaries as predicted by comparison of Fugu and mouse genomic DNA sequence. The similar but not identical position of the second intron and the absence of the fourth intron in the Fugu Surf-6 gene homolog are highlighted. Table 1 indicates the percentage of amino acid identity and similarity of each conceptual Fugu protein to the mouse protein calculated using the GAP program.
Comparison Between the Sizes of the Fugu and Mouse Surfeit Gene Homologs
Table 1 shows a comparison between the sizes of the mouse and Fugu Surfeit gene homologs from initiator codon to termination codon as well as an indication of the difference in intron sizes between each Fugu and mouse Surfeit gene homolog. The genomic distance between the initiator codon and the termination codon of the Fugu Surf-4, Surf-5, and Surf-6 gene homologs is much reduced with respect to the mouse Surf-4, Surf-5, and Surf-6 genes, the Fugu genes being ∼7, 3.5, and 2.5 times smaller, respectively, owing to a sevenfold to twofold reduction in the sizes of their introns. On the other hand, the Fugu Surf-1, Surf-2, and Surf-3/rpL7a gene homologs are of a comparable size with their respective mouse counterparts. The Fugu Surf-1 and Surf-2 gene homologs are both less than twofold smaller than the mouse genes; however, some of the introns are several times smaller in Fuguthan mouse (up to a sevenfold reduction) even though the introns in both species are comparatively small. The Fugu Surf-3/rpL7agene is slightly larger than the mouse Surf-3/rpL7a gene mainly owing to a large first intron in Fugu. The otherSurf-3/rpL7a introns in both species are similar in size and relatively small.
Difference in the Sizes of the Mouse andFugu Surfeit Genes (Including Intron Sizes) and the Amino Acid Homology Between their Predicted Protein Products
Comparison of the Amino Acid Sequence Homology of the Products of the Fugu and Mouse Surfeit Gene Homologs
Table 1 also shows the percentage of amino acid identity and similarity between the predicted Fugu and mouse Surfeit gene polypeptides. It can be seen that the Surf-3/rpL7a, Surf-4,and Surf-5 gene homologs are extremely well conserved at the amino acid level between mouse and Fugu, whereasSurf-2 and Surf-6 are relatively poorly conserved.Surf-1 shows intermediate conservation. It should be noted that cosmids containing the Fugu Surf-1, Surf-2, andSurf-6 gene homologs could not be identified in the initial library screen using mammalian Surf-1, Surf-2, andSurf-6 cDNA probes either because of under-representation of the true cosmid in the well corresponding to the library grid co-ordinate (as was the case for cosmid 186H17) or because the DNA sequence homology between probe and Fugu sequences was too poor under the hybridization conditions used (Surf-2 andSurf-6). Comparison by alignment of amino acid sequences between the Fugu and mammalian Surfeit gene homologs gives an indication of those regions of the proteins that are most likely to be functionally important especially for those that are less well conserved (Fig. 2).
Comparison of the Promoter Regions and Polyadenylation Signals of the Fugu and Mouse Surf-3/rpL7a Gene Hmologs
Usually it is not possible to identify the 5′ end of genes from the analysis of genomic sequences; however, most ribosomal protein genes in vertebrates and some invertebrates possess a polypyrimidine tract at their 5′ end containing the transcriptional start sites. Such a polypyrimidine tract is located at the 5′ end of the Surf-3/rpL7a gene in mammals (Huxley and Fried 1990a; Colombo et al. 1991), birds (Colombo and Fried 1992), and Drosophila(Armes and Fried 1995). Furthermore, the first exon/intron boundary of the mammalian and avian Surf-3/rpL7a genes is demarcated by the splice consensus donor sequence ATG/GT that follows 10–14 bp 3′ of this polypyrimidine tract, the splice occurring directly after the ATG codon that specifies the initiator methionine (Colombo and Fried 1992;Colombo et al. 1991; Huxley and Fried 1990a). With this knowledge we were able to tentatively assign the first exon of the Fugu Surf-3/rpL7a gene by the presence of a polypyrimidine tract 12 bp upstream of a methionine initiator codon and 15 bp upstream of a splice donor site ATG/GT (Fig. 3A).
Characterization of the 5′ and 3′ ends of the Fugu Surf-3/rpL7a gene. (A) Alignment of the Fugu and mouse Surf-3/rpL7a promoter regions. The Fugu and mouse Surf-3/rpL7a gene promoter region sequences were aligned using the GAP program of the GCG software package. Identical nucleotides are indicated by a vertical line. Gaps in the sequence have been inserted to improve the alignment. The codon for the initiator methionine (Met) is boxed and shaded. A polypyrimidine tract in which the mouse gene initiates transcription (Huxley and Fried 1990a) and theFugu gene is suspected to (see text) is boxed as is an upstream element denoted Box B that is known to be conserved in the promoters of the characterized mammalian and avianSurf-3/rpL7a genes. An alignment of the human, mouse, chicken, and Fugu Box B elements generated using the Pileup program of the GCG software package and shown below shows the conservation of this element in more detail with identical bases being boxed and shaded. The position of the Box A element that is conserved between the promoters of the mammalian and avian Surf-3/rpL7a genes is indicated on the mouse sequence by a broken line box. (B) Sequence of the region between the Fugu Surf-3/rpL7a and Surf-1 gene homologs. The sequence is double-stranded, reading 5′ to 3′ on the top strand in the direction of transcription of the Fugu Surf-3/rpL7a gene. The coding sequence contained in the last exon of each gene is boxed. Arrows indicate the direction of transcription of the two genes. Conceptual translation of the coding sequences is shown above the DNA for the Surf-3/rpL7a gene and below the DNA for the Surf-1 gene. Shaded boxes indicate a consensus polyadenylation signal on the top strand used by the Fugu Surf-3/rpL7a gene and two overlapping near consensus polyadenylation sites (5 out of 6 bp) on the bottom strand within the coding region of the Fugu Surf-1 gene, one of which is used by this gene. Bold dots above (Surf-3/rpL7a) and below (Surf-1) the sequence mark known positions of poly(A) tails in cDNA clones isolated to date. The Fugu Surf-3/rpL7a/Surf-1intergenic distance is therefore a minimum of 184 bp compared with 70 bp in mouse.
A number of transcriptional promoter elements have been found to be conserved between the mouse, human, and chicken Surf-3/rpL7agenes (Colombo and Fried 1992). Inspection of the genomic region upstream of the Fugu Surf-3/rpL7a gene has revealed that the Box B element, located just 5′ to the polypyrimidine tract, is also conserved in Fugu, but other promoter elements further upstream, including Box A, are not conserved (Fig. 3A).
Analysis of cosmid 186H17 that contains both the Fugu Surf-3/rpL7a and Surf-1 genes has revealed that the coding regions of these two Fugu genes are separated by only 236 bp and that the genes are convergent. This organization is identical to that found in mouse where the convergent Surf-3/rpL7a andSurf-1 transcription units are separated by only 70 bp (Huxley et al. 1988). The region between the 3′ ends of the Fugu Surf-1 and Surf-3/rpL7a gene homologs is shown in Figure3B. A consensus polyadenylation signal is found 10 bp 3′ to the termination codon of the Fugu Surf-3/rpL7a gene (Fig. 3B). Each of the 14 Fugu Surf-3/rpL7a cDNA clones isolated utilize this polyadenylation signal. Two sites of poly(A) addition downstream of this poly(A) signal have been observed. The position of theFugu polyadenylation signal is comparable with the position of the polyadenylation signals in the mammalian and chickenSurf-3/rpL7a gene homologs relative to the termination codon. There are no consensus polyadenylation signals downstream of theFugu Surf-1 termination codon in this intergenic region, but there are a number of near-consensus polyadenylation signals. Analysis of the three Fugu Surf-1 cDNA clones isolated indicates that one of two overlapping near-consensus polyadenylation signals within the coding region of the gene (4–10 bp 5′ to the termination codon) is used (Fig. 3B). A single site of poly(A) addition is observed in these Surf-1 cDNA clones. Therefore, a minimum of 184 bp separates the 3′ ends of the Surf-1 andSurf-3/rpL7a gene homologs based on the position where their respective poly(A) tails are added. Comparison of the intergenic regions between the Fugu and mouse Surf-3/rpL7a andSurf-1 genes reveals no significant stretches of DNA homology.
Repetitive Elements Are Found in the Sequence 5′ to theFugu Surf-5 Gene Homolog
A cluster of three small (<200 bp) partially inverted sequences are found within the first 1 kb of the 2.5 kb of sequence upstream of the Fugu Surf-5 gene with the first being particularly pronounced. Furthermore, a small sequence element (<300 bp) 1.25 kb upstream of the Surf-5 initiator methionine and positioned next to the inverted sequences is predicted to be a dispersed repetitive element because homologous sequences are found upstream of the Fugu α-anomalous (testis) actin gene homolog (GenBank accession no. U38962). Furthermore, a BLAST search of the Human Genome Mapping Project (HGMP) Resource Center Fugu sequences (http://fugu.hgmp.mrc.ac.uk/) reveals several other Fugucosmid sequences that are homologous to this element. To date, no other repetitive elements have been found in the Fugu DNA we have sequenced.
Base Composition and CpG Methylation Status of the Genomic Regions Containing Fugu Surfeit Gene Homologs
In this study ∼24 kb of Fugu genomic sequence has been obtained in and around the Fugu Surfeit gene homologs. The percentage of guanine plus cytosine (GC) content for this sequence as a whole is 42.6%, and the average relative observed/expected (O/E) CpG dinucleotide frequency predicted by base composition for theFugu genomic sequence we have obtained is 0.61. Similar values for Fugu genomic regions have been reported previously (Elgar 1996; Elgar et al. 1996). The O/E CpG dinucleotide frequency value forFugu (0.61) contrasts with the average CpG O/E frequency of 0.2 for the mammalian genome, the under-representation of the CpG dinucleotide probably being because of the deamination of methylated cytosine in the CpG dinucleotide to thymine. The difference between the CpG O/E frequency of Fugu and mammals suggests a difference in methylation patterns and/or a difference in the number of unmethylated CpG-rich islands per kilobase of genomic DNA. The locus containing theSurf-2, Surf-4, Arginino-Succinate Synthetase (ASS), and EST00098 gene homologs shows extensive relaxation in CpG suppression with an average CpG O/E value of 0.73 for this entire region covering 10 kb, such extensive relaxation not being seen in mammalian genomes. In addition, the same region also shows a percentage GC content (47.2%) significantly higher than the other twoFugu loci containing Surfeit gene homologs (39.6% and 39.0%), although all three values are lower than the value for the human Surfeit locus (53.4%).
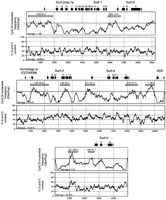
Figure 4 illustrates the base composition and relative CpG dinucleotide frequencies (O/E) for the sequenced regions around the six Fugu Surfeit gene homologs. A significant reduction in CpG suppression and spikes of CpG O/E > 1.0 can be seen at the 5′ end of all of the Surfeit gene homologs and theASS gene homolog (Fig. 4), and a very distinct spike was detected 1.5 kb upstream of the Surf-5 gene that might indicate the presence of another gene in this region although computer searches have not revealed any homology to any sequences in the DNA/protein databases. Further upstream from this point is 1 kb of sequence that shows very high CpG suppression (CpG O/E = 0.13) compared with the rest of the Fugu sequence obtained that corresponds to the location of the repetitive sequences discussed above. Interestingly, the percentage GC content for the threeFugu sequence contigs does not fluctuate greatly, and it can be seen that the regions of reduced CpG suppression are no more GC rich than regions with high CpG suppression.
Base composition and CpG suppression in the three Fugu loci containing Surfeit gene homologs. The three sets of plots show CpG dinucleotide O/E frequencies (calculated by the Staden software suite) and percentage of GC content (calculated by the MacVector 5.0.2 sequence analysis software from The Oxford Molecular Group) for eachFugu locus containing Fugu Surfeit gene homologs. The positions and intron/exon structures of the six complete FuguSurfeit gene homologs are indicated by boxes (exons) and broken lines (introns) above each set of plots. Exon 1 of the ASS gene homolog and the position of the sequences homologous to humanEST00098 are also shown. Arrows indicate the direction of transcription of each gene. Regions have been “sampled” for their CpG O/E value and are marked by shaded bars. The CpG O/E value and the percentage GC content (in brackets) for each sample region is given above the shaded box in each case. Each region has been sequenced in its entirety.
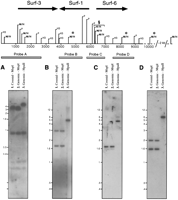
To determine the methylation status of CpG dinucleotides in the promoter regions of three of the Fugu Surfeit gene homologs, we have used the different methylation sensitivities of the restriction enzyme isoschizmers MspI and HpaII. Both enzymes recognize the sequence CCGG; however, HpaII will not cleave the site if the central C (in the CpG dinucleotide) is methylated, whereas MspI is not sensitive to this methylation. The different patterns of DNA migration observed following electrophoresis of MspI and HpaII digests of Fugu genomic DNA on a 1% agarose gel suggest that Fugu genomic DNA is heavily methylated (data not shown). Southern blots of MspI- and HpaII-digested Fugu genomic DNA and anMspI digest of cosmid 186H17 DNA were subsequently probed with radiolabeled restriction fragments spanningMspI–HpaII restriction sites in the promoter and nonpromoter regions of the Fugu Surf-3/rpL7a and Fugu Surf-1/Surf-6 genes to determine any differences in the methylation state of CpG dinucleotides within the MspI–HpaII restriction enzyme recognition sites in this region. Figure5 shows a restriction map of this region showing the predicted MspI–HpaII restriction sites and, below, the results of Southern blot analyses using four different probes (shown labeled A–D) from this region. In Figure 5A probe A hybridizes to three restriction fragments of ∼210, 290, and 900 bp that are common to all three lanes. The probe also hybridizes to an ∼3-kb restriction fragment in the MspI digests of cosmid DNA andFugu genomic DNA (lanes 1,2) but to a larger ∼4.6-kb restriction fragment in the HpaII digest of Fugugenomic DNA (lane 3). This suggests that all threeMspI–HpaII sites spanned by probe A are unmethylated in native Fugu genomic DNA but that the nextMspI–HpaII restriction site along (spanned by probe B) is methylated. Figure 5B shows that probe B hybridizes to ∼1.7- and ∼2.9-kb restriction fragments in MspI digests of cosmid and Fugu genomic DNA (lanes 1,2) but only to a single ∼4.6-kb restriction fragment (as in Fig. 5A, lane 3) in theHpaII digest of Fugu genomic DNA (lane 3). This confirms that the MspI–HpaII site spanned by probe B is methylated in native Fugu genomic DNA. In Figure 5C, hybridization of probe C to two restriction fragments (∼4.6 and ∼6.5 kb) in lane 3 indicates that at least one of theMspI–HpaII sites spanned by probe C is unmethylated in native Fugu genomic DNA because it/they are cleaved in anHpaII digest of Fugu genomic DNA. The ∼4.6-kb fragment is predicted to be the same as that in Figure 5, A and B (lane 3), confirming that the MspI–HpaII site spanned by probe B is methylated in native genomic DNA. The ∼6.5-kb fragment predicts that the MspI–HpaII site spanned by probe D is methylated in native Fugu genomic DNA. Probe C hybridizes to two restriction fragments (∼1.6 and ∼1.7 kb) in theMspI digests of cosmid and Fugu genomic DNA (lanes 1,2) as predicted by the restriction map. In Figure 5D probe D hybridizes to a single restriction fragment of ∼6.5 kb (predicted to be the same fragment as in C, lane 3) in the HpaII digest ofFugu genomic DNA (lane 3) but hybridizes to two restriction fragments (∼1.6 and ∼2 kb) in MspI digests of cosmid andFugu genomic DNA (lanes 1,2) as predicted. HpaII therefore does not cleave the MspI–HpaII site spanned by probe D in digests of Fugu genomic DNA confirming that it is methylated in native genomic DNA. These results also predict that the next MspI–HpaII site 3′ to Surf-6 is also methylated. The analyses therefore indicate that the fourMspI–HpaII sites in the promoter of the Fugu Surf-3/rpL7a gene and at least one site in the promoters of theFugu Surf-1/Surf-6 genes are not methylated, whereas the only site between these two promoters and a site in the 3′ end of theFugu Surf-6 gene and a more 3′ site are methylated (Fig.5). Although we could only test the methylation status of a few CpG dinucleotides using this approach, the results do show that the promoters of the Surf3/rpL7a, Surf-1, and Surf-6genes, which show a CpG frequency predicted by base composition, contain unmethylated CpG dinucleotides, whereas the regions between the promoters contain CpG dinucleotides that are methylated.
Determination of the methylation status of CpG dinucleotides in theFugu locus containing the Fugu Surf-3/rpL7a, Surf-1,and Surf-6 genes by Southern blot analyses. The position ofMspI–HpaII (M/H), PstI (P), andHindIII (H3) restriction enzyme recognition sites are shown along the 9.4 kb of sequence (horizontal line) obtained for the locus containing the Fugu Surf-3/rpL7a, Surf-1, and Surf-6genes. The predicted positions of other MspI–HpaII restriction sites outside the region sequenced are shown above a broken line and are included to facilitate interpretation of the data. The relative positions and orientations of each gene from initiator methionine to termination codon (except Surf-1 whose 5′ end is not determined) are indicated by bold arrows. Probes derived from PstI, HindIII, orPstI–HindIII restriction fragments used in the Southern blot analyses are shown as shaded boxes below and are noted as spanning MspI–HpaII restriction sites. (A–D) A Southern blot analysis probed with the corresponding probe (A–D). For each blot, 0.4 ng of cosmid 186H17 DNA was digested with MspI (lane 1), 3 μg ofFugu genomic DNA was digested with MspI (lane2), and HpaII (lane 3) and run on either a 2% agarose gel (A) or a 1% agarose gel (B,C,D).MspI and HpaII restriction digests of cosmid 186H17 DNA give an identical pattern of restriction fragments indicating that the cloned cosmid DNA contains no MspI–HpaII restriction sites containing methylated CpG dinucleotides (data not shown). Probable methylated CpG dinucleotides withinMspI–HpaII restriction sites as determined by this analysis are indicated by an asterisk (*). (§) At least one of the three MspI–HpaII restriction sites at theSurf-1/Surf-6 promoters is predicted not to be methylated.
DISCUSSION
In this study we have identified homologs of all six of the Surfeit genes in the Japanese puffer fish, F. rubripes. We have shown that the predicted protein products of each gene homolog are well conserved between mammals and Fugu and that the structure of the Fugu genes are very similar, if not identical, to their mammalian counterparts over their coding regions. Only the Fugu Surf-6 gene homolog, which has one fewer intron within its coding region, and the Fugu Surf-1 gene homolog, the 5′ end of which is very poorly conserved, show gene structures that are significantly different to the mammalian genes (introns in noncoding regions were not identified). With the exception of theSurf-3/rpL7a gene (which is slightly larger in Fugu), the Fugu homologs were all found to be smaller than their mouse and human counterparts, but the degree to which the mammalian homologs are expanded in relation to the Fugu homologs differs significantly. The Surf-4 gene shows the greatest difference in size between Fugu and mammals, being about seven times larger in mammals, the Surf-5 and Surf-6 genes show an intermediate size difference, and the mammalian Surf-1 andSurf-2 genes are only moderately expanded when compared with their Fugu homologs. These differences in the degree of expansion of the mammalian genes (or contraction of the Fuguhomologs) may reflect some fundamental difference in DNA turnover for different mammalian genes, a difference that is not manifested so greatly in the Fugu genome. The conservation of gene structure and general reduction in gene size seen in this study supports the potential usefulness of the Fugu genome as a model to predict the genomic structure of mammalian genes.
We were also interested to determine whether regulatory elements might be conserved between the Fugu and mammalian Surfeit gene homologs because conserved regulatory elements have been previously shown to exist between mouse and Fugu in the Hox gene regions (Marshall et al. 1994; Aparicio et al. 1995). However, at present it is not known to what degree housekeeping gene promoters are conserved between these distantly related vertebrates. Significant conservation of promoter elements between the Fugu Surfeit genes and those of higher vertebrates could only be seen for the Surf-3/rpL7agene although shorter stretches of conserved nucleotides were also seen in the Surf-4 and Surf-5 promoters (data not shown). The polypyrimidine tract and a more 5′ conserved element, termed Box B, of the Surf-3/rpL7a gene promoter region are shown to be conserved between mammals, chicken, and Fugu and have enabled us to predict where the 5′ end of the Fugu gene is located (Fig. 3). A more 5′ element that is conserved between mammals and chicken (Box A) is not conserved in Fugu and may therefore only be important for regulation of the Surf-3/rpL7agene when positioned next to the promoter of the Surf-5 gene. A consensus polyadenylation signal can also be seen 10 bp 3′ to the termination codon of the Surf-3/rpL7a gene that is in a conserved position in relation to the mammalian and chicken genes (Fig.3). We have therefore, unusually, been able to predict the 5′ and 3′ ends of the Fugu Surf-3/rpL7a gene based on the positions of conserved regulatory elements. Isolation of Fugu Surf-3/rpL7a cDNA clones confirmed that predictions as to the position of the polyadenylation signal were correct.
Furthermore, we have investigated the promoter regions of the sixFugu Surfeit gene homologs to determine whether they are associated with the presence of CpG-rich islands as is the case with the mammalian and chicken Surfeit gene homologs (Colombo et al. 1992). Our data indicates that all promoter regions of the Fugu genes identified in this study have a reduced suppression of the CpG dinucleotide compared with the nonpromoter regions we have sequenced (Fig. 4). Furthermore, we have shown that at least some of CpG dinucleotides in the promoters of the Fugu Surf-3/rpL7a andSurf-1/Surf-6 genes are unmethylated, whereas CpG dinucleotides within the nonpromoter regions of the same genes are methylated. Both observations suggest that these genes are associated, as are the mammalian gene homologs, with CpG-rich islands; however, whereas mammalian and avian CpG-rich islands are relatively GC rich compared with surrounding DNA, the Fugu CpG-rich islands do not show a raised GC content, a feature consistent with the previously reported characteristic features of CpG islands of other cold-blooded vertebrates (Cross et al. 1991). Although the small Fugugenome may inevitably result in CpG islands comprising a greater percentage of total DNA and affect the average genomic CpG suppression value, a simple calculation suggests that this fact alone cannot account for the difference in CpG suppression values betweenFugu and mammals. Instead, it seems more likely that regions of low CpG suppression must be more widespread in Fugu and that CpG islands may be relatively much larger in the Fugugenome. It is tempting to suggest that there may be a link between the generalized reduction in CpG suppression in the Fugu genome and the small size of its genome. The differences observed in percentage GC content of the three Fugu loci containing Surfeit gene homologs (47.2% for the Surf-4/Surf-2/ASS locus compared with 39.6% and 39.0% for the Surf-3/Surf-1/Surf-6 and Surf-5 loci) do not correlate with differences in GC content within the human Sureit locus but nevertheless suggest that the Fugu Surf-2 andSurf-4 gene homologs are located within a different isochore region of the Fugu genome compared with the otherFugu Surfeit gene homologs.
Mammalian CpG islands are often characterized by numerous consensus Sp1 binding sites, which have often been shown to be important for regulating those genes in transfection studies (Tugores et al. 1994).Fugu genes seem less likely to possess Sp1 sites in their promoters as they are not very GC-rich, which is reinforced by the observation that no Sp1 sites were identified in the promoters of any of the Fugu Surfeit genes in sharp contrast to the situation in mammals. This may indicate that there is a genuine difference in housekeeping gene regulation between cold-blooded and warm-blooded vertebrates or, alternatively, that too much emphasis is often placed on the relevance of Sp1 binding sites in mammals. In this case they may only be frequent because of unusually GC-rich DNA.
Finally, as we are interested in determining the point of origin of the Surfeit locus, it is of some interest to know which particular arrangement of Surfeit genes, either that of the tightly clustered mammalian and avian Surfeit genes or that of the more dispersedFugu Surfeit genes, more accurately reflects the archetypical gene arrangement (Fig. 1). In this respect, it is worth considering what is known of the karyotypic evolution rates found for different vertebrate classes. Studies of karyotypic evolution in fish suggest that the rate of karyotypic change is lower in fish than in birds and mammals (Wilson et al. 1975). Karyotypic changes probably accompany speciation events. Further support for increased karyotypic evolution in birds and mammals as opposed to cold-blooded vertebrates comes from the observation that speciation rate accelerated in birds and mammals compared with cold-blooded vertebrates (Bush et al. 1977; Bernardi 1993). This suggests that Fugu may have a greater propensity to reflect archetypical genomic configurations than mammals and, if true, supports the possibility that the completed Surfeit cluster arose in a restricted fish, amphibian, or reptilian lineage. However,Fugu may not be representative of all cold-blooded vertebrates with respect to the Surfeit locus. Furthermore, Fugu may also have an unusual genomic organization and evolution compared with other teleost fish as its notoriously small genome could reflect different rates of DNA turnover to that of other vertebrates.
To conclude, it is pertinent to address the relevance of these results to the importance of the mammalian Surfeit locus. The Surfeit locus has been demonstrated to be conserved between mammals and birds; however, the existence of an avian Surfeit locus may not necessarily provide a strong case for a requirement for conservation of the locus. The progressive discovery of syntenic regions between mammals and birds (Palmer and Jones 1986; Bumstead et al. 1994; Burt et al. 1995; Li et al. 1995) suggests that the conservation of syntenic regions between birds and mammals may only reflect the relatively slow pace of genomic change in these lineages. With the possible exception of theSurf-3/rpL7a and Surf-1 gene pair, this study has not provided strong circumstantial evidence for a requirement for a conserved order of the Surfeit genes. The organization of the genes in the Surfeit locus may therefore have resulted from random gene shuffling events. It is possible that housekeeping genes are frequently shuffled together because of an increased frequency of breakpoint formation at their promoters resulting from greater DNA fragility in these regions caused by their chromatin status. This evolutionary analysis cannot, however, prove whether coordinate regulation does or does not occur in the Surfeit locus of higher vertebrates.
METHODS
Hybridizations to Libraries and Southern Blots
Southern blotting was performed using the standard protocols suggested in the Hybond-N protocol booklet from Amersham, and Hybond-N membrane was used in all cases. All DNA probes were labeled by random hexanucleotide priming and hybridized with membranes under standard conditions. The Fugu genomic cosmid library used in this study, which is complex enough to cover eight genomes, was obtained from the HGMP Resource Center of the Medical Research Council (MRC). Low-stringency hybridizations to the cosmid libraries and cosmid Southern blots were performed at 55°C, and washes were performed at 58°C in 0.8× SSC (1× SSC is 0.15 m NaCl plus 0.015m sodium citrate), 0.1% sodium dodecyl sulphate (SDS). High-stringency hybridizations of Fugu-derived probes to Southern blots of restriction digests of Fugu genomic and cosmid DNA were performed at 65°C, and washes were at 65°C in 0.1× SSC, 0.1% SDS. Fugu Surf-1 and Surf-3/rpL7agene cDNAs were isolated from a Fugu fish 5′-STRETCH PLUS cDNA library from Clontech using standard high-stringency hybridizations to Fugu probes.
Cloning and Sequencing Techniques
All restriction digests, ligations, and other routine DNA manipulations were performed according to standard protocols, generally as detailed in Sambrook et al. (1989). Sequencing was performed using the Sequenase version 2.0 kit from U.S. Biochemical following the manufacturer’s instructions. Double-stranded sequencing was performed from plasmid DNA minipreparations. Not all sequences were determined on both strands, and ambiguous bases were occasionally encountered, with the exception of the coding regions of the genes where extra care was taken.
Sequence Analysis
All sequences were processed using the MacVector 5.0.2 program from Oxford Molecular Group PLC. This software was also used to calculate and plot the GC content of the sequences and to sample regions for their CpG O/E values. The Genetics Computer Group, Inc. (GCG) software suite was used to generate protein and DNA sequence alignments. The Staden software suite was used to generate the plots of CpG O/E frequency in Figure 4.
Acknowledgments
We thank Drs. Anna-Marie Frischauf and Denise Sheer for their helpful comments in the preparation of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
Sequence analysis of a PCR product derived from the FugucDNA library has demonstrated the existence of the alternatively spliced Fugu Surf-5b mRNA containing a fourth exon (see text and Fig. 2).