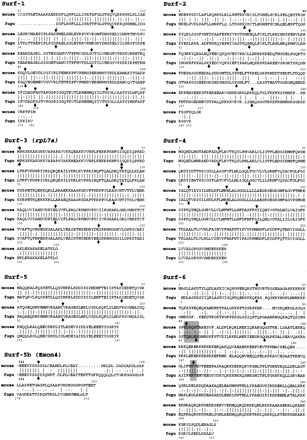
Amino acid homology between the mouse and predicted FuguSurfeit gene products. The GAP program from the GCG software suite was used to generate amino acid sequence alignments comparing each of the six mouse Surfeit proteins with the predicted protein products of the six Fugu Surfeit gene homologs that have been deduced from the predicted structure of each Fugu gene. Vertical lines between the sequences indicate identity, double dots indicate conservation of similar amino acids, and single dots indicate changes found to occur frequently between homologs. Numeration of both proteins is given from their putative initiator methionines except for the putativeFugu Surf-1 protein whose amino terminus has not been elucidated and the Surf-5b protein for which alignment begins at amino acid 131 of both Fugu and mouse proteins. Vertical arrows above the mouse sequences and below the Fugu sequences indicate the relative positions of intron/exon boundaries as predicted by comparison of Fugu and mouse genomic DNA sequence. The similar but not identical position of the second intron and the absence of the fourth intron in the Fugu Surf-6 gene homolog are highlighted. Table 1 indicates the percentage of amino acid identity and similarity of each conceptual Fugu protein to the mouse protein calculated using the GAP program.











