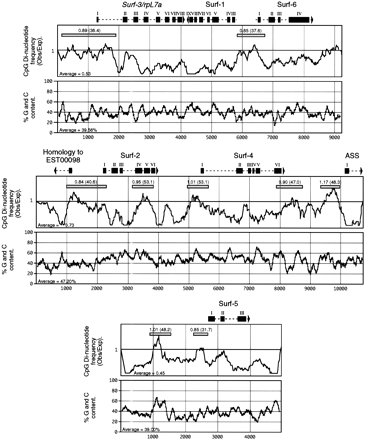
Base composition and CpG suppression in the three Fugu loci containing Surfeit gene homologs. The three sets of plots show CpG dinucleotide O/E frequencies (calculated by the Staden software suite) and percentage of GC content (calculated by the MacVector 5.0.2 sequence analysis software from The Oxford Molecular Group) for eachFugu locus containing Fugu Surfeit gene homologs. The positions and intron/exon structures of the six complete FuguSurfeit gene homologs are indicated by boxes (exons) and broken lines (introns) above each set of plots. Exon 1 of the ASS gene homolog and the position of the sequences homologous to humanEST00098 are also shown. Arrows indicate the direction of transcription of each gene. Regions have been “sampled” for their CpG O/E value and are marked by shaded bars. The CpG O/E value and the percentage GC content (in brackets) for each sample region is given above the shaded box in each case. Each region has been sequenced in its entirety.











