Conserved noncoding sequences provide insights into regulatory sequence and loss of gene expression in maize
- Baoxing Song1,
- Edward S. Buckler1,2,3,
- Hai Wang1,4,
- Yaoyao Wu1,5,
- Evan Rees2,
- Elizabeth A. Kellogg6,
- Daniel J. Gates7,
- Merritt Khaipho-Burch2,
- Peter J. Bradbury3,
- Jeffrey Ross-Ibarra7,8,
- Matthew B. Hufford9 and
- M. Cinta Romay1
- 1Institute for Genomic Diversity, Cornell University, Ithaca, New York 14853, USA;
- 2Section of Plant Breeding and Genetics, Cornell University, Ithaca, New York 14853, USA;
- 3Agricultural Research Service, United States Department of Agriculture, Ithaca, New York 14853, USA;
- 4National Maize Improvement Center, Key Laboratory of Crop Heterosis and Utilization, Joint Laboratory for International Cooperation in Crop Molecular Breeding, China Agricultural University, Beijing 100193, China;
- 5Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518124, China;
- 6Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA;
- 7Department of Evolution and Ecology, University of California Davis, Davis, California 95616, USA;
- 8Center for Population Biology and Genome Center, University of California Davis, Davis, California 95616, USA;
- 9Department of Ecology, Evolution, and Organismal Biology, Iowa State University, Ames, Iowa 50011, USA
Abstract
Thousands of species will be sequenced in the next few years; however, understanding how their genomes work, without an unlimited budget, requires both molecular and novel evolutionary approaches. We developed a sensitive sequence alignment pipeline to identify conserved noncoding sequences (CNSs) in the Andropogoneae tribe (multiple crop species descended from a common ancestor ∼18 million years ago). The Andropogoneae share similar physiology while being tremendously genomically diverse, harboring a broad range of ploidy levels, structural variation, and transposons. These contribute to the potential of Andropogoneae as a powerful system for studying CNSs and are factors we leverage to understand the function of maize CNSs. We found that 86% of CNSs were comprised of annotated features, including introns, UTRs, putative cis-regulatory elements, chromatin loop anchors, noncoding RNA (ncRNA) genes, and several transposable element superfamilies. CNSs were enriched in active regions of DNA replication in the early S phase of the mitotic cell cycle and showed different DNA methylation ratios compared to the genome-wide background. More than half of putative cis-regulatory sequences (identified via other methods) overlapped with CNSs detected in this study. Variants in CNSs were associated with gene expression levels, and CNS absence contributed to loss of gene expression. Furthermore, the evolution of CNSs was associated with the functional diversification of duplicated genes in the context of maize subgenomes. Our results provide a quantitative understanding of the molecular processes governing the evolution of CNSs in maize.
The genomes of a million eukaryote species will likely be sequenced within the next decade (Lewin et al. 2018), but understanding how these genomes work without ENCODE-scale projects and data (The ENCODE Project Consortium 2012) for each species will require that we also use evolutionary approaches to identify key functional regions. In general, noncoding sequences (CNSs) occupy a larger portion of the genome than coding regions. Most genome-wide association hits have been reported to be located in the noncoding regions in, for example, maize and humans, and are enriched in putative gene expression regulatory sequences (Wallace et al. 2014; Zhang and Lupski 2015; Nishizaki and Boyle 2017; Giral et al. 2018). Comparison of noncoding sequences across species can identify regions under purifying selection to reveal functional constraint (Guo and Moose 2003; Vandepoele et al. 2006; Haudry et al. 2013; Finucane et al. 2015; Polychronopoulos et al. 2017; Xiang et al. 2019). However, detection of conserved noncoding sequences in plants is an ongoing challenge (Van de Velde et al. 2016), receiving extensive recent attention in a broad range of species (Inada et al. 2003; Freeling and Subramaniam 2009; Algama et al. 2017; Polychronopoulos et al. 2017; Xie et al. 2018). A genome-wide comparison of features of putative functional elements (Zhang et al. 2012; Rodgers-Melnick et al. 2016; Oka et al. 2017; Wang et al. 2017b; Li et al. 2019; Lu et al. 2019; Ricci et al. 2019; Tu et al. 2020) with CNSs could provide new insight into understudied noncoding fractions of the genome.
Genomes of the grass tribe Andropogoneae provide a valuable and powerful system for the study of conserved sequences. Species of the Andropogoneae tribe have diverged in a relatively short time frame, sharing a common ancestor ∼16–20 million years ago (Vicentini et al. 2008). Andropogoneae species include maize, sorghum, sugarcane, and silvergrass, some of the most productive grain, sugar, and biofuel crops worldwide (Manners 2011; Brosse et al. 2012). Andropogoneae species share the NADP-ME C4 photosynthesis system (Black et al. 1969; Sage and Zhu 2011) and similar development patterns, whereas their genomes are highly diverse with frequent polyploidization (Estep et al. 2014) and extremely active transposable elements (TEs) (Ramachandran et al. 2020). Nevertheless, despite rapid sequence turnover elsewhere in Andropogoneae genomes, functional sequences are expected to be under purifying selection, making the tribe an ideal system in which to identify and understand the role of CNSs.
Results
An atlas of CNSs in the Andropogoneae tribe
Inspired by recent studies of regulatory architecture (Oka et al. 2017; Lu et al. 2019; Ricci et al. 2019; Parvathaneni et al. 2020; Tu et al. 2020), we used coding genes as anchors and developed a sensitive sequence alignment pipeline to identify CNSs in Andropogoneae genomes which have undergone genome-wide duplications, numerous rearrangements, and gene loss (Fig. 1; Schnable et al. 2011). Andropogoneae genomes (see below) were aligned to the maize B73 v4 assembly (Jiao et al. 2017), which was used as a reference. First, we lifted over maize protein-coding genes to each query genome (Fig. 1, step 1) by mapping coding sequences (CDSs) using minimap2 (Li 2018). Gene copy number often varied between the maize genome and the query genome, so all minimap2 mapping hits with a similarity larger than 60% were used as anchors, where similarity is the number of identical base pairs to the CDS length in maize. All CDSs and high-frequency k-mers were then removed from the genomes (Fig. 1, step 2). Next, introns, sequences of 100 kbp upstream of the translation start codon and sequences of 100 kbp downstream from the translation stop codon were extracted to perform pairwise alignment. The selection of a 100-kbp range was based on previous observations that almost all open chromatin regions and transcription factor binding sites (TFBSs) are located within 100 kbp of the nearest gene (Rodgers-Melnick et al. 2016; Ricci et al. 2019; Tu et al. 2020). Pairwise alignment was conducted following the widely used seed-and-extend process (Altschul et al. 1990; Li and Homer 2010). Briefly, the query sequences were cut into fragments with overlapping sliding windows using a window size of 38 bp and a step size of 8 bp. The Smith-Waterman algorithm (Smith and Waterman 1981) was employed to align the fragment in each window against the reference sequence, and any alignments with an alignment score ≥40 were used as seeds (Fig. 1, step 3). Every seed was then extended, and extension was terminated using the X-drop approach (Fig. 1, step 4; Zhang et al. 1998). Alignments with a dynamic programming score ≥54 were defined as CNSs. This score threshold corresponds to a P-value < 0.1 assuming that a pair of sequences with a length of 100 kbp were aligned (Fig. 1, step 5; Karlin and Altschul 1990). Finally, the removed CDS and k-mer sequences were put back into aligned CNSs (Fig. 1, step 6) and the SAM-format alignments were generated (Fig. 1, step 7; Li et al. 2009).
Procedures to identify CNSs in Andropogoneae. The maize B73 v4 genome was used as reference (red lines), whereas the other five genomes were individually used as a query (green lines). First, full-length CDS of each maize protein-coding gene was mapped to the query genome (CDSs belonging to the same gene are linked with “>” in the cartoon) (1); then we deleted CDSs (orange lines) and high-frequency k-mers (blue lines) (2). Next, upstream, intron, and downstream sequences were pairwise aligned using a dynamic programming algorithm (3–4). Candidate fragments below a P-value threshold (0.1) were defined as CNSs (5–7).
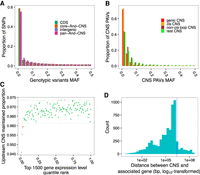
Three publicly available genomes, sorghum (Sorghum bicolor) (McCormick et al. 2018), maiden silvergrass (Miscanthus sinensis) (Zhang et al. 2018), and wild sugarcane (Saccharum spontaneum) (Mitros et al. 2020) were used as queries (see Supplemental Table S1 for more information regarding all the genomes used in this study). In addition, the genomes of two heterozygous Andropogoneae species, Hyparrhenia diplandra and Chrysopogon serrulatus, were assembled to supplement the above-mentioned three genomes (Fig. 2A). Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness scores (Simão et al. 2015) of 0.94 demonstrated that these two newly assembled genomes had high completeness and low sequencing error rates. The median distance between genes and contig edges were 35 kbp and 77 kbp in the genomes of H. diplandra and C. serrulatus, respectively, indicating the contiguity of these two assemblies was suitable for the detection of CNSs within 10 kbp of >85% of coding genes (Supplemental Fig. S1).
Pan-Andropogoneae CNSs. (A) Phylogenetic relationships of Andropogoneae species used in this study. Andropogoneae species are in the green shaded portion of the phylogeny. (B) Simulation of the total length of pan-And-CNSs and core-And-CNSs by iterative random sampling of taxa. Red and blue lines indicate the pan- and core-And-CNS curves fit using points from all combinations.
Total CNS length identified using each query genome was positively correlated with the query genome size (Supplemental Fig. S2). The sorghum genome is the only monoploid assembly in our data set that has not experienced a genome-wide duplication event after its divergence from the common ancestor of Andropogoneae (https://doi.org/10.6084/m9.figshare.1538627.v1). Aligning the genome sequence of sorghum against that of maize generated the smallest CNS space (67.07 Mbp, by counting matched base pairs in the maize genome). The largest CNS space was observed by aligning genome sequences between wild sugarcane and maize (86.97 Mbp). The sizes of CNSs ranged from 27 bp to 15 kbp (Supplemental Fig. S3). The total length of CNSs present in maize and at least one other species was 106.52 Mbp, accounting for ∼5% of the maize genome. Hereafter, those CNSs were designated as pan-Andropogoneae CNSs (pan-And-CNSs). A total of 42.27 Mbp CNSs were present in all species and were termed as core-Andropogoneae CNSs (core-And-CNSs). However, more species are needed to better describe pan- and core-And-CNSs (Fig. 2B).
Pan-And-CNSs were enriched with putative cis-regulatory elements (Supplemental Fig. S4) and overlap with 52%–78% of sequences with putative cis-regulatory features (TFBSs, open chromatin regions, acetylation of histone 3 lysine 9 [H3K9ac] ChIP-seq peaks, micrococcal nuclease hypersensitive regions [MNase HS], or DNase I hypersensitive sites [DHSs]) identified in different plant tissues (Supplemental Table S2). We also confirmed that a few known regions were correctly classified as CNSs: the third intron of knox1 (also known as knotted1) (Supplemental Fig. S5; Greene et al. 1994; Lai et al. 2017), vgt1 (Supplemental Fig. S6; Salvi et al. 2007), and tcp (also known as tb1) (Supplemental Fig. S7; Clark et al. 2006; Studer et al. 2011), suggesting the identification of functional noncoding sequences using our approach.
A large proportion of CNSs overlap with putative regulatory elements
A large proportion (86.8%) of genes have pan-And-CNSs detected within 2 kbp upstream. When compared with genes without pan-And-CNSs detected within 2 kbp upstream, genes with CNSs have higher expression levels and less tissue expression specificity (Yanai et al. 2005; Kadota et al. 2006) across 23 maize B73 tissues (Supplemental Fig. S8; Walley et al. 2016). Fifty-one percent of the pan-And-CNSs overlap with the untranslated region (UTR) or intron of coding genes (herein genic CNS, otherwise intergenic CNS) (Supplemental Fig. S9); this proportion is comparable to that of Arabidopsis thaliana (Haudry et al. 2013). Because introns and UTRs have a wide range of conserved functional roles (e.g., to promote gene expression, guide splicing, produce noncoding RNA) (Greene et al. 1994; Akua et al. 2010; Chorev and Carmel 2012; Ritchie and Flicek 2014; Rigau et al. 2019), we did not classify genic CNSs further by potential function. The overlap between intergenic pan-And-CNSs and low DNA methylation loci (Xu et al. 2020) was higher than the genome-wide random expectation (Supplemental Fig. S10). Fifty-six percent of intergenic pan-And-CNS records overlap with open chromatin regions, TFBSs, or H3K9ac ChIP-seq peaks (Fig. 3A), a 6.5- to 19.5-fold enrichment relative to random expectation (Fig. 3B). Compared to a previously released list of conserved elements in the maize intergenic region (Tian et al. 2020), we generated a larger data set and our intergenic CNSs have greater overlap with open chromatin regions, TFBSs, and H3K9ac ChIP-seq peaks (Supplemental Fig. S11).
CNSs are primarily putative regulatory sequences. (A) Proportions of intergenic pan-And-CNSs overlapping with features of putative cis-regulatory sequence. (B) Enrichment of intergenic pan-And-CNSs in open chromatin regions, TFBSs (transcription factor binding sites), H3K9ac (acetylation of histone 3 lysine 9) ChIP-seq peaks, chromatin loop anchors, TEs (transposable elements), and noncoding RNA (ncRNA) genes.
To further uncover the function of intergenic pan-And-CNSs, we analyzed colocalization of intergenic pan-And-CNSs with chromatin loop anchors, TEs, and noncoding RNA (ncRNA) genes. We started with chromatin loop anchors because they were inferred to be conserved and their dynamics are associated with transcription activity (Dong et al. 2017, 2018, 2020; Harmston et al. 2017; Liu et al. 2017; Polychronopoulos et al. 2017; Wang et al. 2018; Delaneau et al. 2019; Szabo et al. 2019). Chromatin loop anchors identified by Hi-C and HiChIP (Peng et al. 2019; Ricci et al. 2019) overlapped with 47% of intergenic pan-And-CNS records, with an enrichment of 6.6-fold compared to the genome-wide background (Fig. 3B). In addition to open chromatin regions, TFBSs, and H3K9ac ChIP-seq peaks, chromatin loop anchors overlapped with an additional 12% intergenic pan-And-CNS records (Supplemental Fig. S12). Regulatory elements derived from TEs have been described by several independent studies (Xie et al. 2006; Smith et al. 2008; Dupeyron et al. 2019), and cases of TEs acting as regulatory sequence have been reported (Xie et al. 2006; Smith et al. 2008; Studer et al. 2011; Makarevitch et al. 2015; Zhao et al. 2018; Dupeyron et al. 2019; Noshay et al. 2020). Enrichment of CNSs overlapping TEs was not observed (Fig. 3B), but when looking at particular TE superfamilies (Stitzer et al. 2019), 1.08- to 1.78-fold enrichments of intergenic pan-And-CNSs were observed in four of the 13 TE superfamilies (RIL, RST, DHH, and DTM) (Supplemental Fig. S13). However, these four TE superfamilies only accounted for another 2.09% of intergenic pan-And-CNSs. Finally, enrichment of CNSs in ncRNA genes (Han et al. 2019) was also observed (Fig. 3B), which accounted for only 1.04% of the total intergenic pan-And-CNSs.
CNSs have diverse functions
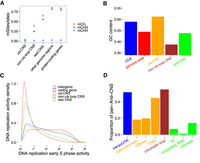
Due to the tissue-specific activity of some cis-regulators, a full data set of putative cis-regulatory sequences has not been generated until the present study. It is essential to know if CNSs that do not overlap with features of cis-regulatory elements might have alternative functions. We therefore classified intergenic CNSs into the following groups: (1) CNSs overlapping with open chromatin, TFBS, or H3K9ac ChIP-seq peaks (cis CNS); (2) CNSs overlapping with chromatin loop anchors not part of the group cis (non-cis loop CNS); and (3) remaining CNSs not included in (1) and (2) (rest CNS). We then investigated the DNA methylation ratio, guanine-cytosine (GC) content, and DNA replication activity in the early S phase of the mitotic cell cycle in each CNS group. Different DNA methylation ratios, GC content, and DNA replication activity in the early S phase were obtained for these CNS groups, indicating these CNSs may have a diversity of functions.
In terms of DNA methylation, the cis pan-And-CNSs showed a low DNA methylation ratio, which supports the previous observation that putative cis-regulatory sequences correspond to a low DNA methylation ratio in plants (Zhang et al. 2006, 2012; Zilberman et al. 2007; Suzuki and Bird 2008; Rodgers-Melnick et al. 2016; Oka et al. 2017; Ricci et al. 2019). The non-cis loop pan-And-CNSs exhibited a medium DNA methylation ratio (Fig. 4A) and could be divided into two distinct subgroups according to DNA methylation ratios (Supplemental Fig. S14). This might be due to different functions of CNSs between these two subgroups; alternatively, some may actually be cis CNSs with tissue specificity but were incorrectly classified into the non-cis loop pan-And-CNS group. DNA methylation is associated with GC content (Mugal et al. 2015) and GC content is associated with chromatin accessibility (Parker et al. 2008; Schwartz et al. 2019; Hammelman et al. 2020). Compared to the genome-wide background, the cis pan-And-CNSs showed a higher GC content, whereas non-cis loop pan-And-CNSs exhibited a lower GC content. This is again suggestive of diverse functions across cis pan-And-CNSs and non-cis loop pan-And-CNSs. The GC content of the rest of the pan-And-CNSs was similar to that of the genome-wide background (Fig. 4B). Active regions of DNA replication in the early S phase (Wear et al. 2017) were associated with both cis pan-And-CNSs and non-cis loop pan-And-CNSs. The activity of the rest pan-And-CNSs was higher than that of the genome-wide background but lower than that of cis pan-And-CNSs and non-cis loop pan-And-CNSs (Fig. 4C; Supplemental Fig. S15).
Patterns of DNA methylation and GC content in CNSs suggest diverse functions. (A) Different DNA methylation ratios among pan-And-CNS groups. Red dots correspond to CG DNA methylation, green dots are CHG DNA methylation, and blue dots represent CHH DNA methylation (where “H” indicates A, C, or T). “other genome regions” on the horizontal axis represents DNA methylation sites located in the intergenic regions that were not defined as CNSs, and “protein-coding genes” denotes DNA methylation sites located within CDSs, introns, or UTR regions of coding genes. (B) Different groups of pan-And-CNSs (indicated in orange, brown, and green) have distinct GC content when compared with CDSs (blue) or the genome-wide (red). (C) Overlap of CDS regions, cis, non-cis loop, and rest pan-And-CNSs with active regions of DNA replication in the early S phase of the mitotic cell cycle. Sequences that did not overlap with coding genes or CNSs were used as background (intergenic). (D) The proportion of pan-And-CNSs overlapping with annotated features. Each CNS can overlap with multiple features. Unknown CNSs are those CNSs that do not overlap with any used features.
Overall, 14% of pan-And-CNS records did not fall in any putative features (i.e., open chromatin regions, TFBSs, introns, UTRs, noncoding RNA genes, chromatin loop anchors, H3K9ac ChIP-seq peaks, and TEs) (Fig. 4D; Supplemental Fig. S16). These CNSs were shorter and had lower alignment scores (Supplemental Fig. S17) than the 86% that could be assigned to a feature and may be enriched for false positives. These unannotated pan-And-CNSs might also be tissue-specific regulators. A more comprehensive investigation of functional noncoding features in different tissues may provide a better understanding of CNS functions.
Variants in CNSs impact gene expression
We further investigated the CNS function by testing if genotypic variants within the identified CNS affected maize gene expression. Using the expression quantitative trait loci (eQTLs) identified by Kremling et al. (2018), the intergenic pan-And-CNSs were enriched 4.02-fold among the “lead” eQTLs compared to the genome-wide background. We selected the lead eQTL as the one with the strongest association (lowest P-value) with the expression of its target gene. The pan-And-CNS regions harbored a larger proportion of maize HapMap3 variants (Bukowski et al. 2018) with low minor allele frequency (MAF) when compared to intergenic regions (Fig. 5A). This result suggests that variants in the CNS regions are under stronger purifying selection compared to those in the intergenic regions, likely because variants in CNSs could negatively impact functional elements.
Variants in CNS regions impact gene expression. (A) MAF distribution of HapMap3 variants in CNS regions, genome-wide CDS regions, and genome-wide intergenic regions. (B) MAF distribution of CNS PAVs in genic, cis, non-cis loop, and rest CNS groups. (C) Comparison of the proportion of maintained CNSs in the 2-kbp upstream regions of the top 1500 expressed genes in root tissues in each maize accession. Dotted lines indicate the 99% one-tailed confidence interval calculated by shuffling the gene expression ranks and CNS maintained proportions 1000 times. Red dots are beyond the 99% one-tailed intervals. Similar patterns were observed across different tissues (Supplemental Fig. S18). (D) Histogram of the distance between CNS PAVs and associated genes for root expression data when a PAV and its associated genes are on the same chromosome. The vertical dotted line indicates a distance of 2.5 Mbp.
We identified CNS presence/absence variants (PAVs) using the maize HapMap3 second-generation sequencing reads (Bukowski et al. 2018) and CNSs identified using the sorghum genome as the query. Most CNS PAVs were rare (MAF < 0.1), especially genic CNSs and cis CNSs (Fig. 5B). Previous studies suggested that noncoding rare variants contribute to the dysregulation of nearby downstream genes and are negatively associated with organism fitness (Flint-Garcia et al. 2005; Kremling et al. 2018). By analyzing the CNS absence within the 2-kbp upstream region of a gene and its expression, we observed loss of CNSs was associated with loss of gene expression (Fig. 5C). However, further upstream, the association between loss of CNSs and loss of gene expression was much weaker (Supplemental Fig. S18); beyond 2 kbp, the regulatory activity of CNSs on downstream genes likely diminishes. To further test the impact of CNS PAVs on gene expression, we used gene expression profiles of seven tissues from a modern inbred population (Kremling et al. 2018) along with the CNS PAVs with MAF ≥ 0.1 to perform an expression genome-wide association study (Supplemental Fig. S19). The result showed that more than half of significant CNS PAVs were located within 2.5 Mbp of associated genes (Fig. 5D; Supplemental Fig. S20).
Finally, we investigated the evolution of CNSs in maize during domestication and subsequent local adaptation. We identified regions likely selected during domestication using genome-wide selective sweeps using RaiSD (Alachiotis and Pavlidis 2018) in a panel of 31 maize landraces (Wang et al. 2017a) and regions important for local adaptation in maize using pcadapt (Luu et al. 2017). The overlap between these regions and pan-And-CNS was substantially less than would be expected by chance, suggesting recent selection in maize has mainly favored variants that do not modify these constrained functional regions (Supplemental Fig. S21).
CNS variation is associated with functional diversity between maize subgenomes
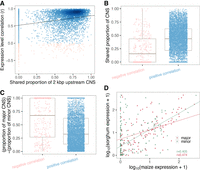
To further study the effect of CNS variation on gene expression, we took advantage of the genome-wide duplication event that occurred in maize after divergence from sorghum (Swigonová et al. 2004) to investigate CNS variation between the two maize subgenomes. Syntenic homologous genes between genomes of maize and sorghum were identified using the quota-alignment implementation (Tang et al. 2011) with parameters that keep every two maize genes corresponding to one sorghum gene (‐‐quota) (see Methods). First, for each orthogroup (including two maize genes and one sorghum gene), the total CNS length within the 2-kbp upstream region of a gene was recorded if this CNS site was present within the 2-kbp region of the sorghum homologous gene and at least one maize homologous gene copy (Supplemental Fig. S22). Second, the proportion of shared CNS sites was calculated as the number of CNS sites present in both maize gene copies to that of the total CNS length. Third, the expression similarity between two maize gene copies was calculated using Pearson's correlation of their expression levels across 23 tissues of maize B73 (Walley et al. 2016). Then, a correlation analysis of the proportion of shared CNS sites and expression similarity was conducted. The proportion of CNS sites shared between the two paralogous genes was positively correlated with the expression similarity between them (r2= 0.10, P-value < 2.2 × 10−16, Pearson's correlation) (Fig. 6A; Supplemental Fig. S23). Moreover, maize paralogs with negatively correlated expression patterns shared a significantly smaller proportion of CNS sites than positively correlated paralogs (P-value < 2 × 10−16, Wilcoxon rank-sum test) (Fig. 6B). Here, we defined the gene copy with a longer CNS (within 2-kbp upstream region) as the major copy and the gene with a shorter CNS as the minor copy. In the context of maize subgenomes, for each pair of maize syntenic paralogous genes with negative expression correlation, in addition to spatiotemporal expression patterns, we also observed a higher nonsynonymous to synonymous mutations ratio for the minor CNS approximation gene (Supplemental Fig. S24), which may indicate neofunctionalization or pseudogenization. The size difference between the major and minor CNSs was smaller in positively correlated paralog pairs than in negatively correlated paralog pairs (P-value = 5 × 10−10, Wilcoxon rank-sum test) (Fig. 6C).
CNS variation is associated with expression diversity between paralogous genes in maize. (A) Correlation of CNS similarity and expression similarity of paralogous gene pairs. Red dots indicate negatively correlated genes; blue dots indicate positively correlated genes across tissues. (B) The shared proportion of CNS sites for negatively (red) and positively (blue) correlated paralogous gene pairs. (C) The diversity of CNS maintained by the maize major copy and minor copy for negatively (red) and positively (blue) correlated gene pairs. (D) Correlation of expression levels of the maize major copy genes with their sorghum homologous genes (red) and minor copy genes with their sorghum homologous genes (green) in shoots for genes with negatively correlated expression patterns across maize tissues in panel A.
In addition, for those negatively correlated paralog pairs, we examined the correlation between the expression of maize genes and their sorghum homologs in the shoot tissues. The normalized shoot RNA-seq data of B73 (maize) (Kremling et al. 2018) and sorghum (NCBI BioProject [https://www.ncbi.nlm.nih.gov/bioproject/] accession number PRJNA503076) were retrieved from Washburn et al. (2019). We observed a higher correlation between the expression levels of maize major copies and the homologous sorghum genes than between maize minor copies and the homologous sorghum genes in the shoot (Fig. 6D). Overall, these results suggest that CNS variation is associated with expression and functional diversity between duplicated genes.
Discussion
Despite the challenges of working on genomes with vast numbers of transposons and frequent duplications, a novel sensitive alignment approach shows that noncoding regions under purifying selection can be identified by comparing genomes of related species. In our CNS identification pipeline, the aim of using coding genes as anchors is to narrow down the sequence alignment scope and reduce false positives. There are some scenarios where our pipeline will fail to identify the target gene. The nearest gene of a regulatory element is not necessarily its target, and the distance between a regulatory element and its target genes might be longer than 100 kbp. In addition, the proximity of a functional noncoding sequence and its target gene may be impacted by large insertions or chromosomal rearrangements. Previous studies suggested that the total number of predicted TFBSs is much smaller than that of transcription factor (TF) recognition sites. TFs often work cooperatively and the sequence contexts of TF recognition sites or combinatorial recognition of cis-elements are key for TF binding specificity (Gerstein et al. 2012; Dror et al. 2015; Levo et al. 2015; O'Malley et al. 2016; Tu et al. 2020; Avsec et al. 2021). CNS records identified in this study are much longer than a single TF recognition site, suggesting the possibility of recognition of ordered combinations of nonoverlapping TFBSs (Viturawong et al. 2013; Shen et al. 2020). CNSs identified using our approach were highly enriched in TFBSs, open chromatin regions, H3K9ac ChIP-seq peaks, chromatin loop anchors, and eQTLs.
Using the genome sequences of six Andropogoneae species, we were able to identify a set of core-And-CNSs and pan-And-CNSs. Core-And-CNSs and pan-And-CNSs showed similar enrichment in potential regulatory sequences, suggesting that not all functional noncoding elements are conserved across all the species. The presence/absence of those elements may be related to species-specific traits (Khalturin et al. 2008; Won et al. 2019). As expected based on the role of core-And-CNS in more essential functions (Cvijović et al. 2018) and the likelihood of mutations in these regions carrying a higher deleterious burden (Kistler et al. 2018), we observed a larger number of variants with low MAF in core-And-CNS regions than in pan-And-CNS regions in a maize population (Fig. 5A), indicating stronger purifying selection.
The overlap between CNSs and features of putative cis-regulatory sequences has been reported previously (Haudry et al. 2013; Viturawong et al. 2013; Warnefors et al. 2016; Lai et al. 2017; Oka et al. 2017; Lu et al. 2019; Ricci et al. 2019), although not all cis-regulatory sequences are evolutionarily conserved (Ross et al. 1994; Wittkopp et al. 2002; McLean et al. 2011). We observed enrichment of lead eQTLs in CNS regions and association between CNS absence and gene expression (Fig. 5C,D). In terms of the regulation of CNSs on target genes, we were able to show that upstream 2 kbp CNS absence was strongly correlated with loss of expression, both in the context of natural variation segregating within a maize population and between duplicated genes in the maize subgenomes. Our results indicate that a longer conserved region proximal to a maize gene correlates with higher expression and similarity of its orthologous gene in sorghum (Fig. 6D), suggesting pseudogenization or neofunctionalization of the gene copy with a shorter CNS. In summary, our study shows that a meta-analysis using CNSs, and gene expression levels combined with open chromatin regions, TFBSs, chromatin loop anchors, and low DNA methylation loci can provide a more comprehensive view of the molecular mechanisms underlying the regulation of gene expression.
Analysis of the identified CNSs extended our knowledge of CNS function. Around half of the identified intergenic CNSs overlapped with different groups of putative cis-regulatory features (e.g., open chromatin regions, TFBSs, and H3K9ac ChIP-seq peaks). Chromatin looping is important for gene regulation (Kadauke and Blobel 2009), and subtle changes in chromatin loop anchors are associated with differential gene regulation and expression (Greenwald et al. 2019; Diehl et al. 2020). GC content and DNA methylation ratio might influence DNA stiffness/flexibility and has been reported to be correlated with DNA supercoiling and recombination (Naughton et al. 2013; Rodgers-Melnick et al. 2015; Jabbari et al. 2019). In this study, we also observed distinguishable GC content and DNA methylation ratios between cis CNSs and non-cis loop CNSs, suggesting diverse functions between these two types of CNSs. ncRNAs can interact with DNAs, RNAs, and proteins, and have been implicated in the regulation of gene transcription and translation, as well as in response to stresses and stimuli (Yoon et al. 2013; Chen and Aravin 2015; Han et al. 2019; Yao et al. 2019; Statello et al. 2021). The enrichment of CNSs in ncRNA genes indicates sequence conservation of transcribed but untranslated DNA sequences. The enrichment of CNSs in active regions of DNA replication in the early S phase suggests that regions of DNA replication initialization might be evolutionarily conserved. CNS knockout lines should be generated to gain a comprehensive understanding of their function (Gasperini et al. 2020). Taking advantage of the observation of GC content and DNA methylation pattern across different groups of CNSs, a model to classify CNSs into different functional groups might be useful to provide a guide for the verification of CNS functions via molecular approaches.
Methods
Plant materials collection
Hyparrhenia diplandra was collected in Kenya by Rémy Pasquet (Pasquet 1126) and Chrysopogon serrulatus was obtained from the USDA Germplasm Repository Information Network (PI 219580; seed originally from Pakistan). Both plants were grown in the greenhouse at the Donald Danforth Plant Science Center. Vouchers of flowering specimens were deposited at the herbarium of the Missouri Botanical Garden; full specimen data are available through the Tropicos database (https://www.tropicos.org).
DNA preparation and sequencing
Total DNA was extracted from young leaf tissues. Long-read sequencing was conducted on a Nanopore MinION platform at the Institute of Biotechnology, Cornell University. DNA with a size of 20–80 kbp was selected following the Blue Pippin protocol, and the selected DNA were cleaned using AMPure XP beads. DNA repair and end-prep were performed with NEB enzyme kits. After adapter ligation, MinKNOW software was used for quality control of the MinION sequencing library. Sequencing was performed following the manufacturer's instructions.
A total of 1.0 μg of DNA per sample was used as input material to generate second-generation sequencing reads. Sequencing libraries were conducted using the NEBNext DNA Library Prep kit following the manufacturer's recommendations, and barcodes were added to each sample. Genomic DNA was randomly fragmented to a size of 350 bp. Then, DNA fragments were end-polished, A-tailed, and ligated with the NEBNext adapter for Illumina sequencing, and further PCR-enriched by P5 and indexed P7 oligos. PCR products were purified (AMPure XP system) and the resulting libraries were analyzed for size distribution by an Agilent 2100 Bioanalyzer and quantified using real-time PCR. The qualified libraries were fed into Illumina NovaSeq sequencers after pooling according to their effective concentration and expected data volume.
Genome assembly
We used the NanoPlot (De Coster et al. 2018) and Porechop package (https://github.com/rrwick/Porechop) to respectively check and filter the MinION raw reads. The MinION clean reads were then assembled using Flye v1.4.2 (Kolmogorov et al. 2019) with the genome size estimated via flow cytometry (Supplemental Fig. S1F) as reference. The MinION clean reads were mapped to the assembly using minimap2 (Li 2018) with a setting of “-x map-ont”, and racon v1.3.1 (Vaser et al. 2017) was used to polish the assembly with default parameters. Assembly polishing using MinION reads was repeated three times. The MEM module of BWA v0.7.17 (Li and Durbin 2009) was used to map Illumina reads to the MinION polished assembly with parameters “-k11 -r10”. The “markdup” command implemented in SAMtools v1.09 (Li et al. 2009) was used to remove duplicated Illumina reads. Pilon v1.23 (Walker et al. 2014) with parameter “‐‐diploid ‐‐fix bases” was used for error correction (Supplemental Fig. S1A). Assembly correction using Illumina reads was repeated three times.
Genome assembly evaluation
To evaluate the contiguities of our new assemblies, we identified 5592 Benchmarking Andropogoneae Single-Copy Orthologs (BASCO) genes shared by four Andropogoneae genomes, maize (Zea mays), sorghum (Sorghum bicolor), maiden silvergrass (Miscanthus sinensis), and wild sugarcane (Saccharum spontaneum), as well as an outgroup species, foxtail millet (Setaria italica). The URLs to access these genomes are listed in Supplemental Table S1.
The following procedures were conducted to identify the BASCO genes:
-
Syntenic genes: CDSs of the B73 genome (v4.34) were first aligned against the sequences of sorghum, wild sugarcane, maiden silvergrass, and foxtail millet using BLASTN (Altschul et al. 1990) with parameters “-outfmt 6 -strand plus -task blastn -evalue 5 -word_size 7 -max_target_seqs 1000”. Next, the quota-alignment pipeline (Tang et al. 2011) was used to detect syntenic genes using parameters “‐‐tandem_Nmax = 5 ‐‐cscore = 0.2 ‐‐no_strip_names ‐‐filter_repeats” for blast_to_raw.py. The parameters of quota_align.py were set as “‐‐merge ‐‐Dm = 20 ‐‐min_size = 3”. In addition, for specific species, the “‐‐quota” parameter was set as “2:2” for maiden silvergrass, “1:2” for foxtail millet, “1:2” for sorghum, and “4:2” for wild sugarcane according to their different genome assembly ploidy or genome-wide duplication levels. Overall, 25,155 maize genes were found in at least one syntenic region.
-
Orthogroups: OrthoFinder (Emms and Kelly 2019) with parameter “-S blast -M msa” was used to find orthogroups. Only those orthogroups with 1–2 maize genes, 1 sorghum gene, 1–2 maiden silvergrass genes, 1–4 wild sugarcane genes, and 1 foxtail millet gene were kept. The CDS of each transcript in the selected orthogroups was mapped to the corresponding genome sequence using minimap2 with parameters “-ax splice -a -uf -C1 ‐‐cs”. Any transcripts with a higher than expected number of hits were removed.
-
Intersect syntenic genes and orthogroups: Orthogroups with at least one syntenic maize gene were kept, otherwise dropped.
-
Double check gene copy numbers: Similar to step 2; CDSs of the genes that passed the previous filter were mapped to the five genomes using minimap2 and filtered by the number of hits within each genome. Orthogroups that passed this last check were defined as BASCO.
CDSs of BASCO genes were mapped to the H. diplandra and C. serrulatus assemblies using minimap2 with parameters “-ax splice -a -uf -C 1 -k 12 -P -t 12 ‐‐cs”. To evaluate the contiguity of the assemblies, we defined the minimum extent of flanking for mapped genes (Supplemental Fig. S1C) as
-
n1 = start position of mapped CDS
-
n2 = contig length – end position of mapped CDSs
-
minimum extent of flanking regions = minimum (n1, n2)
BUSCO v3.1.0 with parameters “-m geno -sp maize -f -r -l” and the liliopsida_odb10 database were used to evaluate the completeness of the assemblies.
Parameter optimization for identification of CNSs
The pipeline started by finding anchor points using minimap2 (Li 2018) with parameters “-x splice -a -uf -C 1 -k 12 -P ‐‐cs”. Keeping in mind the high diversity of the Andropogoneae noncoding regions, for the following alignment steps we used a match score “2”, mis-match “−3”, gap opening penalty “−4”, and gap extension penalty “−2”. For step 3 (Fig. 1), we kept alignments with a minimum Smith-Waterman score of 40 as seeds, thus the minimum seed length was 20 bp (minimum seed score/match score). A sliding window size of 38 bp was selected to ensure there was only one seed in each window. To reduce the computational time and minimize the number of missing seeds, the sliding step size was set as 8.
To identify high-frequency k-mers, we counted the frequency of 20-mers (minimum seed length) using KAT v2.4.2 (Mapleson et al. 2017). For each genome, the secondary derivative of the k-mer frequency density distribution was calculated and the point with minimum distance to zero was identified. The k-mer frequency at that point was used as a threshold to define and remove high-frequency k-mers.
To find the alignment score that corresponds to a P-value <0.1, we randomly extracted 10,000 fragments with a length of 1000 bp from the unmasked reference genome and query genomes separately. A total of 10,000 maximum Smith-Waterman scores were calculated by aligning those fragments, and results were fit into a nonlinear least square regression. The final k (0.006662) and λ (0.382291) values were determined by using maize as the reference against sequences extracted from the other five species randomly. Based on those k and λ values, alignments with a Smith-Waterman score of 54 or higher were kept.
Overlap and enrichment analysis
The output SAM files were reformatted into BAM files using SAMtools, and the “depth” command of SAMtools was used to check
how many unique base pairs were classified as conserved. We counted how many unique CNS base pairs overlapped with open chromatin
regions (Ricci et al. 2019; Tu et al. 2020). Then, enrichment values were calculated as

The overlap and enrichment values for TFBSs (Tu et al. 2020), H3K9ac ChIP-seq peaks (Oka et al. 2017), chromatin loop anchors (Peng et al. 2019; Ricci et al. 2019), TEs (Stitzer et al. 2019), ncRNA genes (Jiao et al. 2017; Han et al. 2019), genome-wide DNA methylation (Ricci et al. 2019), DNA replication profiles (Wear et al. 2017), and lead eQTLs (Kremling et al. 2018) were calculated in the same way.
The above-mentioned features were obtained from the original publications without further processing. For the data sets using the B73 v3 genome assembly coordinates, CrossMap v0.2.8 (Zhao et al. 2014) and the chain file released from Ensembl (Howe et al. 2020) were used to uplift to the maize B73 v4 genome assembly coordinates.
CNSs and other features were considered overlapping if they shared ≥1 bp.
Comparison of gene expression for genes with and without CNS within 2 kbp upstream
A total of 28,950 gene IDs from the expression matrix from 23 maize tissues (Walley et al. 2016) were uniquely lifted from the maize B73 v3 to the v4 genome annotation using the table released by MaizeGDB (gene_model_xref_v4.txt, https://www.maizegdb.org/search/gene/download_gene_xrefs.php?relative=v4, downloaded on April 22, 2019, also available at https://github.com/baoxingsong/dCNS/blob/master/data/gene_model_xref_v4.txt) (Lawrence et al. 2004). Among those genes, 25,127 have upstream CNSs detected within a 2-kbp range. The tissue specificity of gene expression was measured using τ (Yanai et al. 2005) and entropy (Kadota et al. 2006).
CNS PAVs in a maize population
For each accession in the maize Goodman panel (Flint-Garcia et al. 2005), paired-end Illumina reads (NCBI BioProject PRJNA389800) (Bukowski et al. 2018) were mapped to the maize B73 v4 reference genome using BWA-MEM v0.7.13 (Li and Durbin 2009) with default parameters. To save computational time, we used GATK v3.8 (McKenna et al. 2010) to classify each base pair as “callable” or “noncallable.” For the “noncallable” base pairs, coverage was checked using the “samtools mpileup” command. Some CNS regions were shorter than the reads and only a single end of the paired-end reads fell into the conserved region. Therefore, we also mapped the reads to the CNS fragments using BWA-MEM with parameters “-a -c 200000 -S -P” and calculated the coverage of the CNS fragments using “samtools mpileup”. A base pair of a CNS fragment was classified as “present” if it was “callable,” if it had coverage from reads mapping to the genome-wide, or if it had coverage from reads mapping to the CNS fragment; otherwise, it was classified as “absent.” Maize accessions with low sequencing coverage (≤14 Gbp of reads) were excluded from the analysis, as there was not enough information to accurately conduct calling (Supplemental Fig. S25).
Association analysis using CNS PAVs as independent variables
To define CNS PAVs variables, the number of present base pairs/CNS length was calculated for each CNS in each maize accession. If the ratio was ≥0.8, it was encoded as 1; if the ratio was ≤0.2, it was encoded as 0; otherwise unknown. To associate CNS PAVs with gene expression, we used CNS PAVs with a minor allele count ≥15 and a known allele count ≥35. The list of maize accessions analyzed is available as Supplemental Table S3.
Twenty-five PEER factors and three principal components for population structure generated by Kremling et al. (2018) were used as covariants, and association analysis was conducted using a fixed linear model. Association P-values were calculated using an F-test, with a significance threshold 1 × 10−6. The association test was performed using a custom Python script (https://github.com/baoxingsong/dCNS/blob/master/scripts/CnsBasedGwasFixedModelV2.py).
We further investigated the significant associations between CNS PAVs and gene expression level. To further check the possible functional/loss-of-function states of alleles, especially those with a presence ratio <0.8 and >0.2,
-
For each significant association, we grouped those accessions with CNS PAV encoded as 1 as group “presence,” and grouped those accessions with CNS PAV encoded as 0 as group “absence.” For the significantly associated gene, we calculated the median expression level of group “presence” accessions and group “absence” accessions separately.
-
For each of those accessions with the CNS presence ratio <0.8 and >0.2; if its expression level is closer to the group “presence” median value, we classified it into group “presence.” If its expression is closer with group “absence” median value, we classified it into group “absence.”
-
We then compared the CNS reads mapping coverage ratio of group “absence” accessions with group “presence” accessions.
Detection of selective sweeps
We mapped the raw reads from resequenced landraces (Wang et al. 2017a) to the maize B73 v4 reference genome using BWA-MEM (Li and Durbin 2009) with default parameters and conducted SNP calling using the GATK v3.8 (McKenna et al. 2010) germline SNP calling best practices. Specifically, first HaplotypeCaller was used to call variants per sample and create GVCF files. Following this, we used GenomicsDBImport to consolidate GVCF files and joint-called genotypes from these with GenotypeGVFs. SelectVariants was used to output SNPs, as the sweep detection software currently cannot handle indels. We conducted genome-wide scans for selective sweep patterns using RaiSD v2.5 (Alachiotis and Pavlidis 2018) with default parameters, correcting for the number of base pairs with usable sequence data using mop (https://github.com/RILAB/mop) with default parameters.
In order to screen for adaptive loci as a complement to our domestication loci screen, we used the program pcadapt (Luu et al. 2017). We used the program on its default settings but removed inversion Inv4m and known inversions on Chromosomes 1 and 3, as well as a region of low recombination at the end of Chromosome 10 that is likely the abnormal 10 region (Mroczek et al. 2006). We removed these regions as low recombination regions interfere with the way that pcadapt defines the background relatedness for comparisons.
CNS similarity between two maize subgenomes
Syntenic genes between maize and sorghum, introduced above, were used here. To calculate the proportion of CNS sites shared in the 2 kbp upstream of the duplicated genes, we calculated the total length of the sorghum CNSs and the number of matched base pairs shared between each maize gene copy and sorghum gene. Then, we checked the proportion of those shared base pairs that overlapped (Supplemental Fig. S22).
Data access
All the CNS files in SAM format and BED format have been submitted to figshare (https://figshare.com/articles/dataset/CNS/14129006) and are available as Supplemental Data. The CNS identification program source code has been submitted to GitHub (https://github.com/baoxingsong/dCNS) and is available as Supplemental Code. All the sequence reads and de novo assembled genomes generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA543119.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank Maria Katherine Mejia-Guerra for suggestions and comments on the open chromatin and TFBS analysis and William F. Thompson for insights on the DNA replication active regions. We thank Rémy Pasquet for providing plant material of Hyparrhenia diplandra and the USDA Germplasm Repository Information Network for material of Chrysopogon serrulatus. We thank Emre Cimen for help with statistical analysis, Guillaume Ramstein for discussion of eQTL and GWAS analysis, and Qi Sun and Cheng Zhou for advice about the design of sequence alignment algorithms. We also thank Robert Bukowski, Qi Sun, and Arcadio Valdes Franco for sequencing read mapping and variant calling of the maize data. Gen Xu provided suggestions on identification of low-methylated loci. Sara Miller assisted with proofreading the manuscript. We thank the three anonymous reviewers for insightful suggestions and comments. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation #ACI-1548562. This project is supported by the U.S. Department of Agriculture–Agricultural Research Service (USDA-ARS) and National Science Foundation #1822330 to E.S.B., E.A.K., J.R.-I., M.B.H., and M.C.R.
Author contributions: B.S., M.C.R., and E.S.B. designed the experiments and wrote the manuscript. B.S. and E.S.B. implemented the dCNS software. B.S. and Y.W. identified CNS. B.S. and E.R. performed genome assembly. B.S. and H.W. conducted maize subgenome analysis. P.J.B., B.S., and M.K.-B. conducted association analysis. E.A.K. and M.C.R. generated new sequencing data. J.R.-I. and D.J.G. conducted the domestication and local adaptation scan. J.R.-I., E.A.K., and M.B.H. contributed ideas and provided technical help. All authors revised and reviewed the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.266528.120.
- Received May 27, 2020.
- Accepted May 21, 2021.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see https://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.