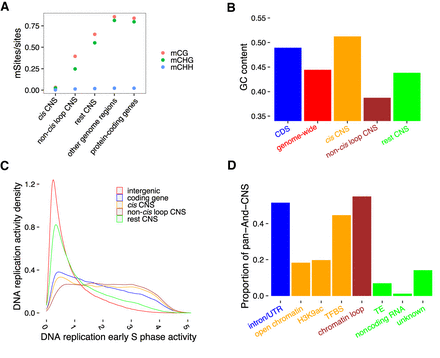
Patterns of DNA methylation and GC content in CNSs suggest diverse functions. (A) Different DNA methylation ratios among pan-And-CNS groups. Red dots correspond to CG DNA methylation, green dots are CHG DNA methylation, and blue dots represent CHH DNA methylation (where “H” indicates A, C, or T). “other genome regions” on the horizontal axis represents DNA methylation sites located in the intergenic regions that were not defined as CNSs, and “protein-coding genes” denotes DNA methylation sites located within CDSs, introns, or UTR regions of coding genes. (B) Different groups of pan-And-CNSs (indicated in orange, brown, and green) have distinct GC content when compared with CDSs (blue) or the genome-wide (red). (C) Overlap of CDS regions, cis, non-cis loop, and rest pan-And-CNSs with active regions of DNA replication in the early S phase of the mitotic cell cycle. Sequences that did not overlap with coding genes or CNSs were used as background (intergenic). (D) The proportion of pan-And-CNSs overlapping with annotated features. Each CNS can overlap with multiple features. Unknown CNSs are those CNSs that do not overlap with any used features.











