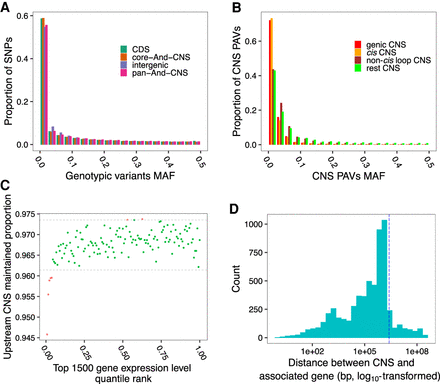
Variants in CNS regions impact gene expression. (A) MAF distribution of HapMap3 variants in CNS regions, genome-wide CDS regions, and genome-wide intergenic regions. (B) MAF distribution of CNS PAVs in genic, cis, non-cis loop, and rest CNS groups. (C) Comparison of the proportion of maintained CNSs in the 2-kbp upstream regions of the top 1500 expressed genes in root tissues in each maize accession. Dotted lines indicate the 99% one-tailed confidence interval calculated by shuffling the gene expression ranks and CNS maintained proportions 1000 times. Red dots are beyond the 99% one-tailed intervals. Similar patterns were observed across different tissues (Supplemental Fig. S18). (D) Histogram of the distance between CNS PAVs and associated genes for root expression data when a PAV and its associated genes are on the same chromosome. The vertical dotted line indicates a distance of 2.5 Mbp.











