Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity
- Nan Cher Yeo1,2,12,
- Caitlin C. O’Meara1,2,12,
- Jason A. Bonomo3,4,
- Kerry N. Veth5,
- Ritu Tomar6,
- Michael J. Flister1,2,
- Iain A. Drummond6,7,
- Donald W. Bowden4,8,
- Barry I. Freedman4,9,
- Jozef Lazar1,10,
- Brian A. Link5 and
- Howard J. Jacob1,2,11
- 1Human and Molecular Genetics Center, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA;
- 2Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA;
- 3Department of Molecular Medicine and Translational Science, Wake Forest School of Medicine, Winston-Salem, North Carolina 27157, USA;
- 4Center for Genomics and Personalized Medicine Research, Wake Forest School of Medicine, Winston-Salem, North Carolina 27157, USA;
- 5Department of Cell Biology, Neurobiology and Anatomy, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA;
- 6Nephrology Division, Massachusetts General Hospital, Charlestown, Massachusetts 02129, USA;
- 7Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA;
- 8Department of Biochemistry, Wake Forest School of Medicine, Winston-Salem, North Carolina 27157, USA;
- 9Department of Internal Medicine - Nephrology, Wake Forest School of Medicine, Winston-Salem, North Carolina 27157, USA;
- 10Department of Dermatology, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA;
- 11Department of Pediatrics, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA
- Corresponding author: jacob{at}mcw.edu
-
↵12 These authors contributed equally to this work.
Abstract
Genome-wide association studies (GWAS) identify regions of the genome correlated with disease risk but are restricted in their ability to identify the underlying causative mechanism(s). Thus, GWAS are useful “roadmaps” that require functional analysis to establish the genetic and mechanistic structure of a particular locus. Unfortunately, direct functional testing in humans is limited, demonstrating the need for complementary approaches. Here we used an integrated approach combining zebrafish, rat, and human data to interrogate the function of an established GWAS locus (SHROOM3) lacking prior functional support for chronic kidney disease (CKD). Congenic mapping and sequence analysis in rats suggested Shroom3 was a strong positional candidate gene. Transferring a 6.1-Mb region containing the wild-type Shroom3 gene significantly improved the kidney glomerular function in FHH (fawn-hooded hypertensive) rat. The wild-type Shroom3 allele, but not the FHH Shroom3 allele, rescued glomerular defects induced by knockdown of endogenous shroom3 in zebrafish, suggesting that the FHH Shroom3 allele is defective and likely contributes to renal injury in the FHH rat. We also show for the first time that variants disrupting the actin-binding domain of SHROOM3 may cause podocyte effacement and impairment of the glomerular filtration barrier.
Genome-wide association studies (GWAS) have identified many genetic loci associated with common complex diseases; however, discovering the exact genes remains problematic for several reasons (Manolio et al. 2009). Furthermore, these loci collectively explain only a small percentage of disease heritability, indicating the need to identify alternative sources of “missing” heritability (Manolio et al. 2009; Chatterjee et al. 2013). Genomic infrastructure available for animal models provides a powerful complementary tool to GWAS (Aitman et al. 2008; Geurts et al. 2009; Cox and Church 2011; Lewis and Tomlinson 2012; Atanur et al. 2013; Varshney et al. 2013). For example, mapping quantitative trait loci (QTL) in mammals and identifying natural mutations in syntenic regions of animal disease models have provided functional support for human risk loci in multiple diseases (Aitman et al. 2008). The ability to then genetically manipulate these disease models enables a direct approach to establish causation (Geurts et al. 2009; Cox and Church 2011). Chronic kidney disease (CKD) is a major health issue and is highly heritable (Jha et al. 2013); however, the majority of risk loci causing CKD are uncharacterized and a vast amount of CKD heritability remains to be explained. Here, we leveraged integrated physiological genomics approaches in zebrafish, rat, and human combined with biochemical structural assessment to dissect a highly replicated GWAS locus (SHROOM3) and its contribution to renal impairment. Based on our study, we propose that similar approaches can be used to assess many of the 44 other uncharacterized human CKD loci and potentially other loci associated with common diseases (Okada et al. 2012; O’Seaghdha and Fox 2012).
Eight GWAS reported association between intronic SHROOM3 variants and CKD (Supplemental Table 1); yet, its role in renal function and disease was unknown, and establishing a functional link between SHROOM3 and renal physiology had been difficult due to early postnatal lethality of Shroom3 knockout in mice (Hildebrand and Soriano 1999). Shroom3 encodes an actin-binding protein that regulates morphogenesis of epithelial cells (Hildebrand and Soriano 1999), and it is expressed in the kidney. We note that Shroom3 is located within a QTL previously found to contribute to albuminuria in the FHH (Fawn-Hooded Hypertensive) rat (Shiozawa et al. 2000). Sequence analysis further identified 13 protein-coding variants in the FHH Shroom3 gene compared to the wild-type allele, implicating it as a potential candidate gene in the FHH rat model of CKD. In the present study, we investigated the role of Shroom3 in kidney function by generating a congenic rat in which the wild-type Shroom3 allele from BN (Brown Norway) rat was introgressed onto the FHH genetic background. We then utilized hypomorphic and tissue-specific disruption of shroom3 in zebrafish to test the effect of the gene and its allelic variants on kidney function. We demonstrate that the variants disrupting the actin-binding domain of SHROOM3 may cause podocyte effacement and impairment of the glomerular filtration barrier. This study establishes the first functional link for SHROOM3 and kidney pathophysiology.
Results
Introgression of the BN Shroom3 gene onto the FHH genetic background improved glomerular function
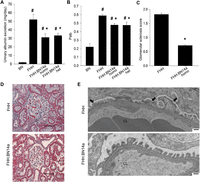
We generated a congenic rat strain (named FHH.BN14a) in which a BN-derived 14.50- to 21.40-Mbp region of Chromosome 14 encompassing the Shroom3 gene was introgressed onto the FHH background and compared its renal phenotypes to the parental FHH strain (Supplemental Fig. 1). At 14 wk of age, both homozygous and heterozygous FHH.BN14a congenic animals showed a significantly lower level of albuminuria, a marker of kidney injury, compared to the FHH rats (Fig. 1A). We measured glomerular permeability to albumin (Palb) in isolated glomeruli from all strains as described previously (O’Meara et al. 2012) and found that Palb in both homozygous and heterozygous FHH.BN14a animals were significantly lower than that in the FHH rats (Fig. 1B). Renal histology showed significantly decreased glomerular sclerosis in the congenic animals compared to the FHH rats (Fig. 1C,D). Electron microscopic analyses of the glomerulus revealed fusion of podocyte foot processes in FHH but not in congenic animals at 18 wk of age (Fig. 1E). These data suggest that replacement of this region containing the BN Shroom3 allele improved glomerular structure and function in the congenic animals.
Introgression of the BN Shroom3 gene onto FHH background improves glomerular and overall kidney function. (A) At 14 wk of age, both homozygous and heterozygous FHH.BN14a congenic animals showed a significantly lower degree of albuminuria compared to FHH (n = 4, 7, 8, and 3, respectively). (B) Both heterozygous and homozygous FHH.BN14a demonstrated significantly improved glomerular permeability (Palb) compared to FHH. (n = 4 animals/115 glomeruli, 3 animals/76 glomeruli, and 2 animals/70 glomeruli, respectively. Palb in BN was obtained from previously published data [Rangel-Filho et al. 2005].) (C) FHH.BN14a kidney showed a decreased presence of glomerular sclerosis compared to FHH at 14 wk of age. A minimum of 30 glomeruli from three kidneys for each strain were scored for a percentage of sclerosis using a scale from 0 (no sclerosis) to 4 (complete sclerosis). (D) Representative trichrome-stained images of glomeruli from FHH and FHH.BN14a are shown. Fibrotic tissues are indicated by blue stain. Scale bars = 50 µm. (E) Electron microscopic images of glomeruli showed podocyte foot process fusion (indicated by arrow) in FHH compared to FHH.BN14a animals at 18 wk of age. Scale bars = 500 µm. (CL) Capillary lumen, (*) P < 0.05 vs. FHH, (#) P < 0.05 vs. BN.
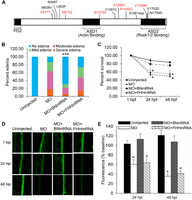
We identified 13 amino acid variants (12 missense and a single-amino acid duplication) in the FHH Shroom3 allele compared with BN rat (Fig. 2A). Six of the 13 variants were predicted computationally to be damaging to Shroom3 function, and several variants localize to conserved functional domains of the protein. Sequence comparison of the entire congenic region identified nine missense variants in addition to those in Shroom3 (Supplemental Table 2). The only other variants predicted to be damaging were located in genes (Stbd1 and Art3) not expressed in the kidney, making Shroom3 the most likely candidate responsible for the observed renal phenotypes in the congenic animals.
FHH Shroom3 is defective and contributes to glomerular dysfunction. (A) Schematic representation of the rat Shroom3 protein is shown. Vertical lines represent the amino acid variants found in FHH Shroom3. Variants predicted to be damaging by PolyPhen-2 are shown in red. (B) Co-injection of shroom3 + tp53 morpholino (MO) with full-length BN, but not FHH, Shroom3 mRNA rescued the edema phenotype. ([***] P < 0.001 vs. MO and MO + FHHmRNA) and (C) cell death induced by shroom3 + tp53 MO ([*] P < 0.05 vs. uninjected control). (D) Representative fluorescence images of individual dorsal aorta at 1, 24, and 48 h following 70-kDa dextran injection are shown. (E) Co-injection with BN but not FHH Shroom3 mRNA rescued the dextran leakage induced by knockdown of endogenous shroom3 in zebrafish. (n = 35, 26, 23, and 22, respectively. [*] P < 0.05 vs. uninjected and MO + BNmRNA.)
FHH Shroom3 failed to restore glomerular function in zebrafish
In situ hybridization detected shroom3 expression in the zebrafish pronephric glomerulus and pronephric tubule (Supplemental Fig. 2). We utilized morpholino (MO)-mediated disruption of shroom3 in zebrafish to assess the impact of shroom3 on kidney function. We injected one- to four-cell stage zebrafish embryos with a MO antisense oligonucleotide that caused skipping of shroom3 exon 5 (Supplemental Fig. 3). A MO against tp53 was co-injected to prevent potential nonspecific cell death (Robu et al. 2007). We observed that uninjected embryos developed normally; whereas shroom3 + tp53 MO-injected embryos predominantly (> 85%) displayed cardiac edema (Supplemental Fig. 4A), consistent with pronephros dysfunction (Kramer-Zucker et al. 2005). A fraction of the shroom3 + tp53 MO-injected embryos also exhibited gastrulation defects. We analyzed the respective groups using an adaptation of a previously described clearance assay to examine pronephric function (Supplemental Fig. 4B; Hentschel et al. 2007), including knockdown of cd2ap, a critical factor for renal glomerular function (Shih et al. 1999), as a positive control in the assay. At 24 and 48 h post-injection (hpi) of 70-kDa fluorescein isothiocyanate (FITC) dextran, we found that the FITC signal was unchanged in the dorsal aorta of uninjected controls and tp53 MO-alone injected zebrafish (Supplemental Fig. 4C,D). In contrast, zebrafish injected with cd2ap + tp53 MO or shroom3 + tp53 MO exhibited significant reduction in FITC signal at 24 and 48 hpi, suggesting that the glomerular size-selective property was compromised after cd2ap or shroom3 knockdown, resulting in an increased rate of clearance of the 70-kDa FITC-dextran (Supplemental Fig. 4C,D).
We next investigated whether co-injection of the zebrafish shroom3 + tp53 MO along with Shroom3 mRNA from either the BN or FHH rats could rescue these defects in zebrafish. Co-injection with BN, but not FHH, Shroom3 mRNA rescued the pericardial edema and gastrulation defects associated with shroom3 + tp53 MO injection (Fig. 2B,C). The glomerular defects induced by knockdown of endogenous shroom3 were also reversed by BN Shroom3 mRNA but not the FHH Shroom3 allele (Fig. 2D,E). These results indicate that the wild-type BN Shroom3 allele is functional; whereas the FHH Shroom3 allele is defective and likely contributes to CKD risk in the FHH rat.
G1073S variant disrupted the actin-binding function of the FHH Shroom3 allele
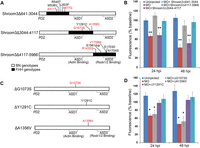
A total of 13 protein-coding variants were identified in the FHH Shroom3 gene, four of which fell in conserved functional domains. We constructed three BN-FHH recombinant Shroom3 cDNAs, where the BN-specific Shroom3 sequence was replaced by FHH alleles from nucleotides 641–3044 bp, 3044–4117 bp, or 4117–5966 bp (Fig. 3A). Co-injection of the shroom3 + tp53 MO with Shroom3∆641–3044 or Shroom3∆4117–5966 mRNA successfully reverted the dextran leakage phenotype observed upon administration of the MO alone; whereas Shroom3∆3044–4117 mRNA did not, suggesting that at least one of the three variants (G1073S, Y1291C, and A1356V) within this region is damaging (Fig. 3B). We then constructed single-variant substitution alleles (i.e., ∆G1073S, ∆Y1291C, and ∆A1356V) and found that the ∆G1073S mRNA failed to rescue glomerular leakage in the zebrafish assay (Fig. 3C,D). The G1073S variant lies in the ASD1 (Apx/Shrm Domain 1) that mediates SHROOM3 interaction with actin (Hildebrand and Soriano 1999), raising the possibility that this aspect of protein function is affected.
The G1073S variant disrupts the function of the FHH Shroom3 gene. (A) Schematic of the different recombinant Shroom3 cDNAs, where a specific region of the BN Shroom3 sequence was replaced by FHH. (B) Co-injection of shroom3 + tp53 MO with Shroom3∆641–3044 or Shroom3∆4117–5966 mRNA, but not Shroom3∆3044–4117 mRNA, rescued dextran leakage induced by the MO (n = 23, 9, 18, 28, and 15, respectively). (C) Schematic of Shroom3 single-amino acid mutants created by site-directed mutagenesis. (D) Co-injection of ∆Y1291C or ∆A1356V restored normal glomerular permeability, while ∆G1073S failed to exhibit functional rescue (n = 17, 12, 17, 23, and 21, respectively). (*) P < 0.05, (**) P < 0.001 vs. uninjected.
Coimmunoprecipitation (co-IP) of a Flag-tagged Shroom3 allele in HEK293 cells demonstrated that the FHH Shroom3 allele exhibited significantly lower affinity for actin than the BN Shroom3 allele, and this defect was recapitulated by the ∆G1073S mutant allele (Fig. 4A–C). The Shroom3 ASD2 domain mediates direct interaction with ROCK proteins (Nishimura and Takeichi 2008), which we examined using the co-IP assay. As expected, mutations in the ASD1 actin-binding domain had no effect on SHROOM3-ROCK1 interaction (Fig. 4A–C). These data demonstrate that the G1073S variant in Shroom3 may disrupt the normal glomerular filtration barrier by compromising the interaction of SHROOM3 with actin.
The G1073S variant decreases the actin-binding affinity of SHROOM3 protein. (A) FLAG-tagged BN, FHH, or ∆G1073S SHROOM3 proteins were overexpressed in HEK293 cells, followed by immunoprecipitation against FLAG. Immunoprecipitated lysates were immunoblotted using antibodies against FLAG, ROCK1, and ACTB. A representative Western blot of immunoprecipitated lysate is provided. (B) Quantification of the Western blot showed that FHH SHROOM3 and ∆G1073S mutant had significantly reduced actin-binding affinity compared to BN SHROOM3. (n = 3 per group. [*]P < 0.05 vs. BN.) (C) ROCK1-binding affinity was not different among the three alleles (n = 3 per group).
P1244L variant disrupted the function of human SHROOM3 gene
GWAS have detected common noncoding variants in SHROOM3 that are significantly associated with CKD, but the overall contribution of these variants to CKD risk is modest (Boger and Heid 2011). To identify other potential contributors to the disease risk, we tested variants in the human SHROOM3 exon5, which harbors the ASD1 actin-binding domain, for association with nondiabetic end-stage kidney disease (ESKD) in 2465 African Americans. The clinical characteristics of the study participants and identification of sequence variants were described in Supplemental Table 3 and Methods. Based on criteria previously described (Bonomo et al. 2014), we selected 20 coding variants and three splice-site variants in the SHROOM3 exon 5 for genotyping.
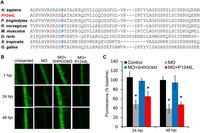
Among the coding variants, rs181194611 showed association with ESKD (P = 0.014, OR = 7.95, MAF = 0.0009) (Supplemental Table 4). This rare variant causes an amino acid substitution from a highly conserved proline to leucine at position 1244 (p.P1244L) (Fig. 5A). P1244L lies downstream from the SHROOM3 ASD1 domain and maps to a homologous region in the mouse ortholog that is known to mediate actin interaction (Hildebrand and Soriano 1999). This variant was predicted to be functionally damaging by PolyPhen-2 (Adzhubei et al. 2010), SIFT (Ng and Henikoff 2001), and PROVEAN (Choi et al. 2012). Co-injection of shroom3 + tp53 MO with the nonmutated human SHROOM3 mRNA restored glomerular function in zebrafish; whereas ∆P1244L mRNA did not (Fig. 5B,C), demonstrating that the P1244L variant indeed impairs SHROOM3 function and is a damaging mutation that is likely contributing to the underlying pathogenesis of glomerular dysfunction.
rs181194611 (p.P1244L) associated with nondiabetic ESKD impairs SHROOM3 function in vivo. (A) Proline at the amino acid position 1244 in SHROOM3 is evolutionarily conserved. (B) Representative images of dorsal aorta at 1, 24, and 48 h following 70-kDa dextran injection are shown. (C) Co-injection of nonmutated human SHROOM3 mRNA restored normal glomerular permeability, while ∆P1244L failed to show functional rescue. (*) P < 0.05 vs. uninjected.
To examine the potential role of noncoding variants in SHROOM3, we tested 11 SHROOM3 noncoding variants previously associated with diabetic kidney disease in African Americans (McDonough et al. 2011) and eight noncoding variants located in transcription factor binding sites. Two common intronic SNPs (rs17002091 and rs17002201) previously identified in diabetic kidney disease exhibited association in this study (P = 0.037 and 0.015, respectively, OR = 0.83 for both SNPs) (Supplemental Table 4). No correlation was found between the two SNPs; thus they are likely distinct signals (r2 = 0.004, D′ = 0.286). Analysis of the rs17002091 and rs17002201 haplotypes identified potentially damaging variants (rs72868158 and rs55650799, respectively), which were predicted to disrupt transcription factor binding sites by RegulomeDB (http://www.regulomedb.org/) (Boyle et al. 2012). Testing of rs72868158 and rs72663250 (proxy of rs55650799) replicated the association previously observed, suggesting that these\ variants underlie the risk associated at this locus. Our results suggest that both common noncoding variants and rare coding variants affecting SHROOM3 expression or function may be contributing to the risk of developing complex kidney disease in humans.
Podocyte-specific disruption of shroom3 caused podocyte effacement in zebrafish
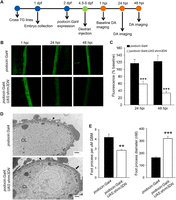
Injuries targeting the glomerular epithelial cells (podocytes) are a major cause of renal impairment in the FHH rat (Simons et al. 1993; Kriz et al. 1998) and proteinuric kidney diseases in humans (Faul et al. 2007; Brinkkoetter et al. 2013). Considering that Shroom3 is expressed in the podocytes (Saleem et al. 2008; Brunskill et al. 2011), we hypothesized that it regulates glomerular permeability via its action on podocyte structure. We generated a zebrafish transgenic line carrying a UAS:shroom3 dominant negative (shrm3DN) allele and crossed it with a line expressing GAL4 under the control of the podocyte-specific podocin (nphs2) promoter (Supplemental Fig. 5). The shrm3DN allele, encoding amino acids 808–1143 of the zebrafish shroom3, inhibited endogenous shroom3 activity (Clark et al. 2012). This knockdown line is a useful tool for dissecting the cell type-specific role of shroom3 and bypasses the confounding effect of global shroom3 knockdown. We assessed the glomerular filtration barrier integrity using a dextran clearance assay shown in Figure 6A. Compared to the podocin:Gal4 control, the podocin:Gal4;UAS:shrm3DN animals showed significantly decreased fluorescence at 24 and 48 hpi, indicating impaired glomerular size-selectivity (Fig. 6B,C). Electron microscopic analyses of the pronephric glomerulus revealed podocyte effacement, indicated by a reduced number of podocyte foot processes and increased foot process width (suggestive of podocyte fusion), in podocin:Gal4;UAS:shrm3DN animals (Fig. 6D,E). These results imply that shroom3 is essential for maintaining the glomerular filtration barrier specifically by regulating podocyte integrity.
Podocyte-specific disruption of shroom3 causes increased glomerular permeability and podocyte effacement in zebrafish. (A) Specific transgenic (TG) lines were crossed to obtain embryos expressing podocin:Gal4 and podocin:Gal4;UAS:shrm3DN. The control and mutants were injected with 70-kDa FITC-labeled dextran at 4.5–5 d post-fertilization (dpf) and analyzed for dextran clearance at 24 and 48 h post-injection (hpi). (DA) Dorsal aorta. (B) Representative fluorescence images of individual dorsal aorta at 1, 24, and 48 hpi are shown. (C) podocin:Gal4;UAS:shrm3DN mutants had significantly decreased FITC signal at 24 and 48 hpi compared to control, indicating that the glomerular filtration barrier was disrupted. (n = 20 and 25, respectively. [***] P < 0.001.) (D) Electron micrograph revealed intact podocyte foot processes (indicated by arrowhead) in podocin:Gal4 animals. Foot process effacement (indicated by arrow) was observed in the podocin:Gal4;UAS:shrm3DN mutants. Scale bars = 500 µm. (CL) Capillary lumen. (E) podocin:Gal4;UAS:shrm3DN had a significantly reduced number of foot processes contacting glomerular basement membrane (GBM) and increased foot process diameter compared to the control. (n = 3 per group. [**] P = 0.01, [***] P = 0.0008.)
Discussion
Mutations targeting the podocyte actin cytoskeleton are the primary cause of many glomerular and proteinuric diseases of the kidney (Faul et al. 2007). The FHH Shroom3 allele with a defective actin-binding domain failed to maintain glomerular integrity in the zebrafish, while the FHH Shroom3 construct containing the functional BN-derived ASD1 domain improved overall kidney function in shroom3-deficient zebrafish. These data suggest that hypomorphic SHROOM3 alleles (e.g., FHH and ∆P1244L) with altered actin-binding likely affect the mechanical characteristics of the glomerular podocyte by disrupting normal actin organization or assembly. It is worth noting that at this point, we do not know whether these alleles are truly sufficient or whether a combinatorial effect of alleles within the haplotype might also exert an effect (Flister et al. 2013).
The actin-binding function of Shroom3 has been shown to be critical for proper morphogenesis of several epithelial tissues (Hildebrand and Soriano 1999; Haigo et al. 2003; Chung et al. 2010; Plageman et al. 2010). For example, Shroom3 regulates apical constriction in neuroepithelial cells by recruiting existing actin fibers to the apical site of the cell (Hildebrand and Soriano 1999). Depletion of Shroom3 in the neural plate prevents recruitment of actin, resulting in loss of the apically constricted morphology without affecting the cell polarity (Hildebrand and Soriano 1999). Thereby, it is possible that Shroom3 regulates the function of kidney podocyte by acting as a morphogenetic regulator. Future studies will be needed to more clearly define the role of Shroom3 in podocytes and investigate whether Shroom3 regulates podocyte cytoskeletons through the same mechanisms observed in other epithelial cell types. Shroom3 interacts with a number of other proteins in addition to actin (Lee et al. 2007; Nishimura and Takeichi 2008). Identification and characterization of these interactions in podocytes will be critical to gain a better understanding of how Shroom3 affects the glomerular filtration barrier.
Using congenic rats, we showed that a locus containing the Shroom3 gene modifies the risk of developing renal impairments. The congenic region contains 60 known and 43 predicted genes, some of which may also affect renal phenotypes. Based on sequence data, Shroom3 appears to be the most likely candidate gene responsible for the renal phenotypes observed in the congenic animals. Transgenic rescue targeting Shroom3 and then correcting the particular variants identified are logical future studies.
FHH rat is a well-established mammalian genetic model of CKD. This particular strain spontaneously develops proteinuria and glomerular injury associated with podocyte effacement (Kriz et al. 1998). Studies using the FHH rat have led to the discovery of genes including Shroom3 that may also be important for kidney diseases in human (Lazar et al. 2013; Rangel-Filho et al. 2013). Sequence comparison between the FHH rat and the renal disease-resistant BN rat identified protein-coding variants in genes (Umod, Alms1, Tfdp2, Dab2, Prkag2, Ino80, Casp9, Mecom, Gnas, and Aldh2), in addition to Shroom3, which have been identified by human GWAS for CKD (Supplemental Table 5), making these interesting candidates for future studies.
Human GWAS have identified several intronic variants in the SHROOM3 gene significantly associated with renal functional traits (Supplemental Table 1); yet, the biological role of the gene/variants has been unclear. The presented studies provide the first functional support for SHROOM3 contributing to renal diseases in humans rather than just being a genetic marker that is correlated with the disease. Follow-up functional investigations in humans, such as fine mapping of SHROOM3, analysis of the gene expression or function associated with the risk haplotype, and case-control studies to test SHROOM3 and the associated LD region, are necessary. Several lines of evidence have suggested that rare variants can play a role in human complex diseases and could account for a significant portion of the disease heritability (Pritchard 2001; Ji et al. 2008; Zuk et al. 2014). Our association analyses identified a rare variant (P1244L) in SHROOM3 that impaired the protein function, suggesting that some rare variants in SHROOM3 may be contributing to renal disease.
In summary, our study shows that Shroom3 is essential for the maintenance of the glomerular filtration barrier specifically by regulating podocyte integrity. We demonstrate that Shroom3-mediated actin interaction is crucial for maintaining normal glomerular function, and mutations within the Shroom3 actin-associating region may modify the risk for renal impairments. Using a combination of strategies across multiple species and data sets, we demonstrate that a rare variant of SHROOM3 is unable to rescue renal function in zebrafish, suggesting that it could play a role in the pathogenesis of glomerular impairment in human disease. We also demonstrate that both common noncoding variants and rare coding variants affecting SHROOM3 expression or function may be contributing to the risk of developing complex kidney disease in humans.
Methods
In situ hybridization
Zebrafish shroom3 cDNA was PCR-amplified using a designed primer pair (Supplemental Table 6) and cloned into the pGEM-T Easy vector (Promega). The plasmid templates were linearized using SacII flanking the 5′-end of shroom3 sequence. Linearized plasmids were subsequently used for in vitro transcription by SP6 RNA polymerase and DIG RNA labeling mix (Roche) to generate the shroom3 antisense probe. Whole-mount in situ hybridization was performed as previously described (Thisse and Thisse 2008) on 48-hpf wild-type zebrafish embryos.
Morpholino and mRNA injections
Morpholino (MO) antisense oligonucleotides were purchased from Gene Tools. The shroom3 MO was designed to target the intron splice-acceptor site upstream of shroom3 exon 5. The effect of the cd2ap MO was previously reported (Hentschel et al. 2007). A tp53 MO was co-injected to inhibit potential nonspecific cell death caused upon injection of the experimental MO (Robu et al. 2007). One hundred micromoles of each MO, diluted in Danieu’s buffer and 0.1% phenol red, were injected in 9.2 nl per one- to four-cell stage zebrafish embryo. To assess the MO-induced splicing defects, total RNA was extracted from MO-injected or uninjected embryos with TRIzol reagent (Invitrogen), and cDNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (Fermentas). PCR was performed over the splice site to detect alternatively spliced products. The alternatively spliced PCR product was gel-extracted and sequenced. To synthesize the mRNA for the rescue experiments, PCR-amplified full-length BN or FHH Shroom3 cDNA was cloned into the pGEM-T Easy vector (Promega). Plasmids were linearized by NcoI and in vitro transcribed into mRNA using the mMESSAGE mMACHINE kit (Ambion) according to the manufacturer’s instructions. Four hundred picograms of either BN or FHH Shroom3 mRNA were co-injected along with the MO into one- to four-cell stage embryos. See Supplemental Table 6 for the sequence of MO and PCR primers.
Zebrafish transgenic lines
The following lines were crossed to make the podocyte-specific shroom3 dominant negative (shrm3DN) mutant line: Tg(podocin:Gal4VP16), and Tg(dnshroom3:UAS:mCherry,myl7:eGFP)mw47. To generate the Tg(podocin:Gal4VP16) transgenic zebrafish, a 5197-nt fragment of the zebrafish podocin promoter was originally cloned from BAC zC116P9 using forward 5′-TGCTACACCATTAAGGTGACCTGTG-3′ and reverse 5′-TCTGTTGTGAAGTGTCCTCT GGTG-3′ PCR primers. A 3.5-kb subfragment was generated using forward 5′-CGGTCACCGGAAGTTTATAAGTATATGGG-3′ and reverse 5′-TCTGTTGTGAAGTGTCC TCTGGTGTTTGG-3′ PCR primers and cloned into pENTER 5′-TOPO vector (Invitrogen) and confirmed by sequencing. The final expression construct, pDestTOl2CG2;podocin:Gal4VP16:polyA, was made using the multisite gateway cloning method (Kwan et al. 2007), and transgenics were generated by plasmid co-injection with Tol2 mRNA into one-cell stage zebrafish embryos. Entry clone plasmids for dnshroom3:UAS:mCherry; myl7:eGFP were made using Gateway technology (Invitrogen) and Tol2-kit reagents (Kwan et al. 2007). Transgenesis of the Tol2 constructs was achieved by co-injection of transposase mRNA into one-cell stage embryos. Details of the dnshroom3 mode of action and generation of the entry plasmids have been described (Haigo et al. 2003; Lee et al. 2007; Clark et al. 2012). The following primers were used to amplify the dnshroom3 fragment: forward 5′-ATGCGATGTGTAAGTCCTGA-3′ and reverse 5′-TCAACTGTATATAAGCACTT-3′.
Zebrafish dextran clearance assay
The dextran clearance assay was adapted and modified from a previous study (Hentschel et al. 2007). For the morpholino and mRNA rescue study, 55-h post-fertilization zebrafish were used for injection of the 70-kDa FITC-labeled dextran. Zebrafish 4.5- to 5-d post-fertilization were used for injection of the dextran in the podocyte-specific shroom3 knockdown study. Experimental zebrafish were anesthetized with tricane and embedded in 1% agarose prior to dextran injections. A total of 9.2 nl of 70-kDa dextran (1 mg/mL) was injected into the zebrafish cardinal vein. Following injection, the dorsal aorta was imaged by confocal microscopy at 1, 24, and 48 h post-injection (hpi). FITC fluorescent intensities were quantified using ImageJ software (National Institutes of Health). The 70-kDa dextran shows a low rate of filtration by an intact glomerulus and therefore should largely remain in circulation. Diminished FITC signal in the circulation at 24 or 48 hpi indicates glomerular leakage.
Transmission electron microscopy (TEM)
Zebrafish were prepared for TEM imaging as previously described (Soules and Link 2005). Briefly, the blocks were trimmed on a Leica RM2255 microtome, and ultrathin sections were cut and collected on coated grids and stained with uranyl acetate and lead citrate. Montaged images of entire cross-sectioned podocytes were captured on a Hitachi H600 at 8000×. Morphometrics of podocytes, number and size of foot processes, were scored in a masked manner where the individual scoring was unaware of the sample genotype.
Rat strain sequencing
FHH/Eur/Mcwi and BN/NHsdMcwi genomic DNA was sequenced using an Illumina HiSeq 2000 and analyzed by CASAVA version 1.8.1 (Illumina). All genomic sequences have been deposited in the NCBI BioProject under accession number PRJEB1333 (Atanur et al. 2013). Variants were analyzed by Variant Effect Predictor (http://www.ensembl.org/info/docs/tools/vep/index.html) (McLaren et al. 2010) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al. 2010).
Generation of FHH.BN14a congenic rat and phenotyping for renal function
The FHH-14BN/Mcwi consomic strain was initially derived from the FHH/Eur/Mcwi and BN/NHsdMcwi inbred strains using marker-assisted selective breeding (Mattson et al. 2007). The FHH.BN14a congenic rat strain (official name: FHH.BN-[D14Rat98-D14Hmgc4]/Hmgc; RGD ID: 8553187) was generated by back-crossing FHH-14BN/Mcwi consomic males to FHH/Eur/Mcwi females. Thirteen- to 14-wk-old male rats were phenotyped for 24-h urinary albumin excretion and in vitro glomerular permeability using methods described previously (O’Meara et al. 2012). After urine collection for the albuminuria study, the left kidney of the experimental animals was collected and immediately fixed with 10% buffered formalin for histological analysis. Fixed tissues were stained and analyzed for glomerular sclerotic injury as previously described (O’Meara et al. 2012). For electron microscopic analysis, kidney cortex from 18-wk-old male rats was collected, prepared, and analyzed as previously described (Rangel-Filho et al. 2013). All animals were housed at the Biomedical Resource Center of the Medical College of Wisconsin (MCW) and maintained on Laboratory Rodent Diet 5001 (PMI Nutrition International Inc.) throughout the studies. Experimental protocols were approved by the MCW Institutional Animal Care and Use Committee.
Cloning of human and rat Shroom3 plasmids
The full-length BN or FHH rat Shroom3 cDNA was amplified using PCR and cloned into a pGEM-T Easy vector (Promega) using the manufacturer’s protocol. BN-FHH recombinant Shroom3 plasmids were generated by cloning with restriction enzymes. The pGEM-T Easy vector containing the full-length BN or FHH Shroom3 cDNA was digested with two enzymes, AgeI and AgrAI, AgrAI and BstEII, or BstEII and NcoI (New England Biolabs). Desired plasmid fragments were gel-extracted and subsequently ligated using T4 ligase (New England Biolabs) to create Shroom3∆641–3044, Shroom3∆3044–4117, and Shroom3∆4117–5966 plasmids (number indicates the base pairs of cDNA replaced by FHH alleles). To create the single amino acid substitution mutant allele, the full-length BN Shroom3 cloned into the pGEM-T Easy vector was PCR-amplified using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Scientific) and phosphorylated primers harboring the mutated sequence. PCR products were subsequently isolated and ligated using T4 ligase. To test the human alleles, we used the commercially available myc-ddk-tagged human SHROOM3 cDNA ORF (Origene), and the mutant allele was generated using site-directed mutagenesis. See Supplemental Table 6 for the sequence of PCR primers used in this study. All plasmids were sequenced and verified.
Protein coimmunoprecipitation (Co-IP) assay
Full-length BN Shroom3, full-length FHH Shroom3, or ∆G1073S mutant cDNA were cloned into the p3xFLAG-CMV-7.1 expression vector (Sigma) and transfected into human embryonic kidney (HEK) 293 cells using a calcium phosphate transfection method. HEK293 cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 1% L-glutamine, penicillin streptomycin (1×), and sodium pyruvate (1×). At 24 h post-transfection, cells were collected and lysed in lysis buffer (200 mM NaCl, 20 mM Tris, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS) to extract protein. Seven hundred and fifty micrograms of protein was used for immunoprecipitation (IP) against FLAG. IP protein products were separated on 4%–20% Bio-Rad mini-PROTEAN TGX gels and transferred onto PVDF membranes, followed by overnight incubation with primary antibodies against FLAG, ACTB, and ROCK1. Membranes were then incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies and developed with enhanced chemiluminescence reagent (Pierce). Protein bands were visualized using a ChemiDoc XRS+ Imaging System (Bio-Rad). Band intensities were quantified with ImageJ software (National Institutes of Health).
Antibodies
Primary antibodies used in this study were: mouse anti-Flag (Sigma, F3165), mouse anti-beta-actin (Abcam, ab6276), and mouse anti-Rock1 (Santa Cruz Biotech, sc-17794). Secondary antibodies used were Peroxidase-AffiniPure Donkey Anti-Mouse IgG (Jackson ImmunoResearch Laboratories, 715-035-150).
Human variant association study
Coding variants in SHROOM3 exon 5 were identified from 1000 Genomes Project and exome sequencing of an admixed African American population (Bonomo et al. 2014). Coding variants in and near the SHROOM3 ASD1 domain were selected for genotyping using criteria previously published (Bonomo et al. 2014). Common noncoding variants in SHROOM3 that showed prior evidence of association (McDonough et al. 2011) and promoter variants predicted to disrupt transcription factor binding by RegulomeDB (http://regulome.stanford.edu/) were also selected for genotyping. Genotyping was performed using the Sequenom MassARRAY platform (Sequenom). Genotype data were analyzed using single SNP association testing as previously described. A P-value of 0.05 was used as the threshold to define significance given the a priori of association between SHROOM3 and measures of renal dysfunction. Details on sample collection, genotyping, and data analyses were previously published (Bonomo et al. 2014). This study was approved by the Institutional Review Board at Wake Forest School of Medicine.
Statistical analysis
Statistical analyses were performed using Sigma Plot 12.0 software. Data are presented as mean ± SEM. All data were analyzed by either one-way ANOVA followed by the Holm-Sidak multiple comparison test (for multiple groups) or unpaired Student’s t-test (for two groups). Dextran clearance results obtained from the morpholino-induced knockdown and mRNA rescue failed the normality test; therefore, the data were analyzed by the nonparametric Kruskal-Wallis test followed by the Dunn’s multiple comparison. The dextran clearance data obtained from podocyte-specific Shroom3 mutant at 48 hpi failed the normality test; therefore, data were log-transformed before t-test analysis. Zebrafish edema incidence was analyzed by the χ2 test.
Data access
The rat sequence variants can be downloaded from the Rat Genome Database in the Genome Browser (http://rgd.mcw.edu/fgb2/gbrowse/rgd_904/) and the Variant Visualizer (http://rgd.mcw.edu/rgdweb/front/select.html).
Acknowledgments
We thank M. Hoffman, C. Purman, M. Cliff, A. Buzzell, A. Lemke, J. Wendt-Andrae, and B. Schilling for excellent technical support; C. Wells for help with electron microscopic imaging; and NIH grant support RO1HL069321 (H.J.J.), RO1-DK071041 (I.A.D.), RO1-DK53591 (D.W.B.), RO1-DK070941 (B.I.F.), and RO1-DK071891 (B.I.F.). J.A.B. is supported by F30 DK098836. This project was conceived and directed by H.J.J.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.182881.114.
- Received August 13, 2014.
- Accepted September 29, 2014.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.