Genomic characterization of the Bacillus cereus sensu lato species: Backdrop to the evolution of Bacillus anthracis
- Michael E. Zwick1,2,6,
- Sandeep J. Joseph3,6,
- Xavier Didelot4,
- Peter E. Chen2,7,
- Kimberly A. Bishop-Lilly2,
- Andrew C. Stewart2,8,
- Kristin Willner2,9,
- Nichole Nolan2,
- Shannon Lentz2,
- Maureen K. Thomason2,10,11,
- Shanmuga Sozhamannan2,
- Alfred J. Mateczun2,
- Lei Du5,12 and
- Timothy D. Read1,2,3,13
- 1Department of Human Genetics, Emory University School of Medicine, Atlanta, Georgia 30322, USA;
- 2Biological Defense Research Directorate, Naval Medical Research Center, Silver Spring, Maryland 20910, USA;
- 3Division of Infectious Diseases, Emory University School of Medicine, Atlanta, Georgia 30322, USA;
- 4Department of Statistics, University of Oxford, Oxford OX1 3TG, United Kingdom;
- 5454 Life Sciences, Inc., Branford, Connecticut 06405, USA
-
↵6 These authors contributed equally to this work.
Abstract
The key genes required for Bacillus anthracis to cause anthrax have been acquired recently by horizontal gene transfer. To understand the genetic background for the evolution of B. anthracis virulence, we obtained high-redundancy genome sequences of 45 strains of the Bacillus cereus sensu lato (s.l.) species that were chosen for their genetic diversity within the species based on the existing multilocus sequence typing scheme. From the resulting data, we called more than 324,000 new genes representing more than 12,333 new gene families for this group. The core genome size for the B. cereus s.l. group was ∼1750 genes, with another 2150 genes found in almost every genome constituting the extended core. There was a paucity of genes specific and conserved in any clade. We found no evidence of recent large-scale gene loss in B. anthracis or for unusual accumulation of nonsynonymous DNA substitutions in the chromosome; however, several B. cereus genomes isolated from soil and not previously associated with human disease were degraded to various degrees. Although B. anthracis has undergone an ecological shift within the species, its chromosome does not appear to be exceptional on a macroscopic scale compared with close relatives.
Bacillus anthracis, the etiological agent of anthrax, has evolved within a branch of the bacterial phylogeny that contains few other mammalian pathogens. Members of the Bacillus genus are Gram-positive endospore-forming bacteria. They are ubiquitous in many environments because they can exploit a wide range of organic and inorganic compounds in the vegetative state and use a dormant spore form to persist through times of starvation and stress. However, although Bacillus is a very diverse genus with more than a hundred species, only the Bacillus cereus group of species is associated with nonopportunistic infections of mammals. This group, comprising B. cereus, Bacillus thuringiensis, B. anthracis, Bacillus mycoides, Bacillus pseudomycoides, and Bacillus weihenstephanensis, is referred to as B. cereus sensu lato (s.l., meaning “in the widest sense”) (Helgason et al. 2000; Jensen et al. 2003; Tourasse et al. 2006). Despite the multiple species names, which are often attributed to phenotypes conferred by mobile genetic elements, all these organisms can be considered members of a single species, because of their low genetic diversity, as measured by 16S sequencing (Daffonchio et al. 2003) and multilocus sequence typing (MLST) (Priest et al. 2004), and their high degree of shared gene content (Rasko et al. 2005). Aside from B. anthracis, other reported virulent strains include a small number of nontraditional anthrax isolates (Hoffmaster et al. 2004; Klee et al. 2010), bacteria that cause wound or soft-tissue infections, a clonal complex of emetic toxin producers (Rasko et al. 2007), and isolates responsible for food poisoning. Most B. cereus s.l. strains isolated have not been linked to mammalian pathogenesis but, instead, are either insect-killing B. thuringiensis or are simply termed “environmental.”
Anthrax is an acute toxemia caused by B. anthracis outgrowth following germination of endospores in its mammalian host. Endospores may enter the host via skin abrasions, ingestion, or inhalation into the lungs. B. anthracis requires expression of a tripartite protein-lethal toxin and a poly-D glutamate capsule for anthrax pathogenesis. These key ingredients in the hyper-virulence of B. anthracis have been acquired through horizontal gene transfer (HGT). The genes encoding the toxin and capsule are on the large plasmids pXO1 and pXO2, respectively. B. cereus s.l. strains are highly variable in plasmid content, suggesting frequent exchange of genetic information via HGT (Jensen et al. 2003; Kolstø et al. 2009). pXO1, pXO2 and plasmids with similar genetic backbones have been found in other Bacillus strains (Pannucci et al. 2002; Rasko et al. 2004). It was therefore predictable that other B. cereus s.l. strains would be found to be associated with anthrax-like disease-containing plasmids strikingly similar to pXO1 and pXO2. B. cereus G9241 caused fulminant pneumonia, with features resembling anthrax in a small cluster of cases from welders in the southern United States in 1987 (Miller et al. 1997). The genome sequence (Rasko et al. 2004) revealed a plasmid almost identical to pXO1 alongside another large plasmid that expresses both exopolysaccharide and hyaluronic acid capsules (Oh et al. 2011). Another recently sequenced strain, “B. cereus var. anthracis” CI, was part of a group of closely related isolates that caused deadly anthrax-like infections in primates in the Côte d'Ivoire in 2001–2002 and Cameroon in 2004 (Klee et al. 2006). The CI strain contained plasmids nearly identical to pXO1 and pXO2 (Klee et al. 2010). Interestingly, B. cereus G9241 and CI belong to a clade of B. cereus s.l. strains that are closely related to B. anthracis.
The origins of virulent anthrax-like strains from multiple nonpathogenic ancestors offers an opportunity to better understand the origins of pathogenicity in this Bacillus group. At one extreme, we may suppose that pathogens in this group have high virulence potential, but their origin is limited solely by the chance event of the HGT of toxin containing plasmids. These types of pathogens have been referred to as “hopeful monsters” (Keim and Wagner 2009). Under this scenario, B. anthracis was such a hopeful monster that seized its opportunity recently and spread clonally around the globe (Kolstø et al. 2009). Alternatively, we might imagine that multiple historical origins of pathogenic strains have occurred because of existing pre-adaptations or newly arising adaptive changes in the genomes of nonpathogenic ancestors. If true, this model predicts that genomic variants will be fixed in the B. anthracis lineage, conferring distinctive phenotypic properties that define the pathogenic niche. In the context of anthrax, taxon-specific genes may help explain why all three pXO1-containing strains have emerged only from one clade. It is thus important to understand whether B. anthracis is unique in its virulence or whether B. cereus G9241, CI, and other pXO1-containing B. cereus s.l. strains we may encounter might be capable of the same level of pathogenicity.
In order to differentiate between these hypotheses, we have used next-generation sequencing to conduct an extensive survey of the genomes of 45 diverse pathogenic and nonpathogenic B. cereus s.l. strains, developing a genome-based phylogeny and examining the pan-genome of the species. Next we tested whether B. anthracis, G9241, CI, or strains in the same clade were unique among B. cereus s.l. in the gain or loss of specific genes (other than the pXO1 plasmid), or had DNA signatures suggestive of a newly emerged pathogen. Instead of finding strong signals in highly pathogenic strains, we uncovered little evidence for adaptive changes in the B. anthracis genome that uniquely predispose it for a virulent lifestyle.
Results
Generation of a phylogenetically representative whole-genome shotgun data set for the B. cereus s.l. species
Forty-five B. cereus s.l. strains were selected for whole-genome shotgun (WGS) sequencing based on geographical, phenotypic, and phylogenetic diversity (Table 1A). For the analysis described subsequently, the 45 WGS sequences were combined with 13 publicly available complete B. cereus s.l. genome projects (Table 1B). To avoid over-representing B. anthracis, which has undergone a recent clonal expansion (Zwick et al. 2005; Van Ert et al. 2007; Kenefic et al. 2009), we chose to include only one genome sequence from this species, the canonical Ames ancestor strain (Ravel et al. 2009). We used the locus tag suffix of the genomes (Table 1A,B) as a naming code to simplify descriptions in this manuscript.
(A) Summary of genomes sequenced
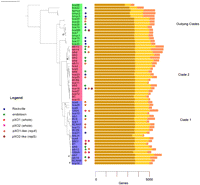
Phylogenetic inference using a distance-based approach that used concatenated chromosomal core proteins from clusters present in all 58 genomes resulted in a tree (Fig. 1) with an almost identical topology to trees produced using concatenated colinear chromosomal segments from a whole-genome alignment of all 58 strains (Supplemental Figs. 1–3). In agreement with earlier studies (Helgason et al. 2000), we found that B. cereus, B. mycoides, and B. thuringiensis strains were not confined within discrete clades and are therefore not monophyletic species. The whole-genome phylogenies of B. cereus agreed well with those from previous studies based on MLST (Priest et al. 2004; Tourasse and Kolstø 2008), and we grouped the strains into clades, using the naming scheme of the first published B. cereus MLST description (Table 2; Priest et al. 2004). Clade 1 contained B. anthracis, as well as several previously sequenced pathogens and other strains linked to virulence (G924 [Hoffmaster et al. 2004], BACI [Klee et al. 2010], BALH [Challacombe et al. 2007], and BCZK [Rasko et al. 2005]). The phylogeny confirmed that the pXO1 plasmid had been acquired on three separate occasions within clade 1 (Klee et al. 2010). Interestingly, this tree revealed that the clade 1 strains most closely related to B. anthracis (Fig. 2) included B. thuringiensis strains isolated from soils in Mexico, Pakistan, and Spain, not known to have any pathogenicity for mammals (serovars monterrey, pulsiensis, and andalousensis, respectively). Serovar monterrey (bthur007) was recently identified as containing poly-D glutamate capsule genes orthologous to B. anthracis (Cachat et al. 2008). Other close relatives of B. anthracis included two human clinical strains that caused endocarditis and fatal pneumonia (bcer16/BCAH8). Serovar konkukian (BT) (Hernandez et al. 1998), which also falls into this “anthracis” lineage, was isolated from a landmine victim in the former Yugoslavia. Clade 2 included emetic toxin-producing pathogens (Ehling-Schulz et al. 2004), as well as numerous environmental B. cereus and B. thuringiensis, and the B. cereus type strain (BC). At the base of the tree was a polyphyletic group of at least three clades, designated “outlying clades” for comparison purposes in this study, containing mostly environmental isolates and more diverse than clades 1 and 2.
B. cereus s.l. summary statistics
Whole-genome phylogeny of B. cereus s.l. The phylogenic tree was constructed from a data set of concatenated, conserved protein sequences using the neighbor-joining (NJ) algorithm implemented by PHYLIP (Felsenstein 1989). The tree was rooted using the known outlier B. cereus subsp cytotoxis. The scale measures number of substitutions per residue. Tree topologies computed using maximum likelihood and parsimony estimates are identical with each other and the NJ tree (see Supplemental Data). We performed 1000 bootstrap replications to test the topology of the tree, and all branches were supported by >50% of the trials. All branches with a bootstrap value of >0.9 are labeled with the following codes: 0.8–0.9; “$”; 0.7–0.8, “#”; 0.6–0.7 “@”; 0.5–0.6 “*”. If not labeled, support was >0.9. Labels refer to the genome codes listed in Table 1. Clades 1 and 2 and the outliers are labeled with blue, red, and green strips, respectively. Circles are opposite genomes containing whole pXO1 (red), pXO2 (fuchsia), or δ-endotoxin genes (green) or were isolated on the same day from Rockville, Maryland (blue). We have also indicated genomes likely containing pXO1-like (brown) or pXO2-like (burgundy) plasmids based on the presence of the RepX (Anand et al. 2008) or RepS (Tinsley et al. 2004) proteins, respectively. Each genome has a bar graph showing the proportion of genes belonging to core (brown), character (ocher), and accessory (pink) classes. All unfinished genomes sequenced in this project can be readily identified because they have three-letter lowercase species identifiers. The only other unfinished genome is G9241. Figures 1 and 2 were drawn using online Interactive Tree of Life (iTOL) software (Letunic and Bork 2007).
Close up of clade 1, rooted at bce9. (Circles) Proportion of orthologous genes present in each genome of the following classes: (red) B. anthracis plasmids, phages, and genetic islands (maximum = 495); (green) all other B. anthracis genes (maximum = 4907); (blue) genes found in other strains, not B. anthracis (maximum = 1580). Bootstrap values of branches over 1000 trials were 1.0 unless indicated. For legend, see Figure 1.
We analyzed patterns of homologous recombination in the 2.74 Mb common to all 58 B. cereus s.l. genomes using ClonalFrame, a software program that determines whether genetic variants arose as de novo mutations or recombination events based on the phylogenetic context (Didelot and Falush 2007). The ratio of the effect of homologous recombination and mutation (r/m) in all B. cereus s.l. strains was estimated to be 2.91 (similar to an earlier estimate of 2.41 based on 13 genomes) (Didelot et al. 2009), which is an intermediate level compared to the extremes of recombinogenic species Neisseria meningitidis and clonal Staphylococcus aureus (r/m of 13.6 and 0.2, respectively) (Didelot et al. 2009; Vos and Didelot 2009). Similar to the results in the earlier study, the r/m estimate for clade 3 was significantly higher than for clades 1 and 2 (4.33 vs. 2.12 and 1.91, respectively) (Table 2). The ClonalFrame analysis reconstructed several hundred recombination events in the terminal branches of the phylogeny leading to each of the three pXO1-containing strains (Supplemental Fig. 3). In the case of B. anthracis, where there are multiple genome sequences available, we could test whether putative recombination events with other B. cereus occurred after the radiation from the common ancestor. In 19 publicly available B. anthracis genomes. we found identical nucleotide sequences at 318 recombinant loci identified by our ClonalFrame analysis, suggesting that the acquisition of the virulence plasmid may have occurred very recently in the B. anthracis lineage, predated by all homologous recombination inferred on the branch above. This argues that in the case of B. anthracis, at least, adaptation to the anthrax-causing niche after the acquisition of pXO1 has not been facilitated through recombination.
Paucity of definitive clade-specific genes in the B. cereus species
We investigated the genetic content among phylogenetic groups of B. cereus s.l. by clustering predicted proteins based on their similarity according to BLAST (Methods). The genome fluidity statistic (Φ; the average pairwise gene content differences between strains) (Kislyuk et al. 2011) is a useful measure of genic variation within a species (Table 2). The value of Φ was 0.22 for all B. cereus s.l. genomes, (meaning that on average any two strains shared 78% of their genes), placed the species between highly cosmopolitan taxa, such as N. meningitidis and Escherichia coli (Φ > 0.3), and the more restricted species, such as S. aureus (0.15) (Kislyuk et al. 2011). Values of Φ calculated for the clade 1 and clade 2 genomes only were both 0.17, significantly lower than the 0.25 for clade 3 (P < 0.05), reflecting the greater phylogenetic diversity of clade 3. Thus, the extent of lateral gene transfer in B. cereus s.l., like homologous recombination, appears to be an intermediate level for a bacterial species.
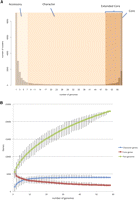
The distribution of the number of genomes in which the 22,975 gene clusters were found was bimodal (Fig. 3A), a similar pattern to that seen in other bacterial species (Holt et al. 2008; Touchon et al. 2009). Based on the approximate inflection points of the U-shaped curve, we defined gene families found in fewer than six genomes as “accessory” and those found in more than 49 as the “extended core” of the species. The genes between these two extremes were termed character genes, using the terminology of Lapierre and Gogarten (2009). While the core defines the essential conserved functions of a species, the character and accessory genes potentially give insight into strain- and clade-specific attributes. Many virulence associated genes fall into these latter classes (Lapierre and Gogarten 2009). The accessory genome (66% total) encoded mostly hypothetical or phage-specific functions. The discovery of accessory genes was still without asymptote after sampling 58 genomes (Fig. 3B), suggesting a large, mobile genetic pool in the species. Although genes of known function were rare, we found novel orthologs of lethal factor component genes pag, lef, and cya in a number of newly sequenced genomes, including environmental strains and an operon of homologs of the TccC and TcaCBA toxin genes of Photorhabdus and Yersinia (Waterfield et al. 2007) in bthur0013 (bthur0013_57400-57440). The finding of novel lethal factor family genes suggested that homologs in other B. cereus s.l. strains may play a role in virulence outside of anthrax, possibly in nonmammalian hosts. The genes encoding Tcc and Tca may have been exchanged during co-infection of insect hosts by Yersinia and B. thuringiensis. The character genes (17% of the pan-genome) constituted were enriched for Gene Ontology (GO) (The Gene Ontology Consortium 2000) terms associated with accessory metabolic and niche-specific survival functions, such as hydrolases, capsule polysaccharide biosynthesis, and beta-lactam antibiotic metabolism (GO:001678, GO:0045227, GO:0030653) (see Supplemental Fig. 4). Many of these genes are located on mobile plasmids, for example, the genes on the backbone of pXO1-like plasmids (Rasko et al. 2007) and the Bacillus thuringiensis δ-endotoxins. The rarefaction curve of character gene discovery (Fig. 3B) suggested that the character gene component of B. cereus s.l. has been completely sampled, with discovery of new genes saturated after about 30 randomly selected genomes.
B. cereus pan-genome. (A) Distribution of gene families across B. cereus s.l. genomes The graph of the number of protein clusters present in B. cereus s.l. genomes. Based on the classification of Lapierre and Gogarten (2009), we defined the extended core as genes encoding proteins present in 49 or more genomes. Accessory genes were present in less than six genomes. The class between these extremes defined the character gene set. The core found in every B. cereus s.l. genome comprised 1754 genes (8% of the total gene clusters). There were a further 2148 genes present in the total extended core of 3904 (17% of the total). These genes may be part of the core excluded by the gene-calling software or sequencing errors in one or more WGS genomes, or were lost in nodes of the B. cereus phylogeny undergoing genome reduction (such as the cytotoxic outgroup strain bce98) (Lapidus et al. 2008). These figures for the core and pan-genome size concur with early estimates by Lapidus et al. (2008) and Han et al. (2006). (B) Rarefaction of pan-genome, character, and core genome estimates. The pan-genome and core genome plots (Tettelin et al. 2005, 2008) were based on protein clustering by Ortho-MCL (Methods). The number of gene families present in the pan-genome or core for n number of genomes was calculated based on 100 trials of genomes inputted in random order. Each point of the median size of the set bars represents maximum and minimum values.
If a branch within the B. cereus s.l. phylogeny were pre-adapted to cause anthrax upon acquisition of pXO1, we might expect to find a stable set of genes exclusively associated with that clade. Instead, we found that very few, if any, genes could be used to define any one clade of B. cereus s.l. When the distribution of accessory and character genes was mapped, clade 1 and 2 were found to have more than 4000 specific gene families (i.e., not found in any other B. cereus s.l. clade) (Fig. 4). However, the number of genes found in all numbers of the clade declined exponentially. This pattern was also held when the subclade of 13 clade 1 genomes containing BACI and GBAA was analyzed (Fig. 4; Supplemental Tables 1, 2; Supplemental Data 1). Aside from pXO1 genes, there are no other genes found only in GBAA, G9241, and BACI (Klee et al. 2006, 2010). This last finding is consistent with the “hopeful monster” model and is not that expected for the model of preadaptation/adaptation of virulent strains.
(A–C). Clade-specific character and accessory genes. Each chart plots the number of clade-specific character present in n or fewer genomes. There are no clade-specific genes present in the 13 genomes that constitute a subclade of close B. anthracis relatives (A; BCZK, bce21, BALH, bce4, BACI, BT, bth9, bth12, GBAA, bth7, bth10, BACH8, bce16) (see Fig. 2) or in all clade 1 strains (B) and only one in clade 2 (C). There are also no clade-specific gene present in all the outlying clade strains (not shown).
Reconstruction of gene turnover using the BadiRate program (Librado et al. 2012) suggested that many character genes had been frequently gained and lost across multiple branches of the B. cereus s.l. phylogeny (Supplemental Fig. 5). Frequent inter-clade exchange was suggested by following patterns of recent insertion sequence (IS) transfers (Supplemental Fig. 6). To visualize how these events may affect the genetic structure of B. cereus s.l., a Neighbor-Net (Huson and Kloepper 2005) network was created based on the presence or absence of all 22,975 genes (Fig. 5). This approach was inclusive of chromosomal genes and genes on plasmids and other horizontally transferred elements. Even though the topology was slightly different than the core protein phylogeny (Fig. 1), because of large-scale gene transfer events (e.g., B. anthracis and B. cereus CI are more closely related on the network because of their shared possession of pXO1 and pXO2) (Klee et al. 2010), the network supported the assignment of each strain to the same clades. There was greater reticulation of the network between clade 1 and clade 2, hinting at a significant level of ancestral gene transfer. The fact that this tree preserves the overall phylogenetic signal of the protein and DNA phylogeny showed that the clades could be defined by their composition of noncore genes and that polyphyletic genes are approximately evenly distributed between strains.
Network phylogeny of the B. cereus s.l. pan-genome. The network was created using the Neighbor-Net algorithm (Kloepper and Huson 2008) based on a binary matrix of protein cluster presence and absence generated by Ortho-MCL. Splits in the network indicate the possibility of recombination between branches.
Clade-specific nucleotide selection patterns
We investigated the possibility that there may be differential patterns of selection acting on orthologous genes, reflecting different ecological pressures on the genomes. The PAML (Yang 2007) likelihood ratio test (LRT) was used to test for positive selection. However, in the manner implemented here, it was dependent on fitting individual gene histories on a whole-genome phylogeny and was thus restricted to the core genes. With the proviso that screens for positive selection can have multiple interpretations (Kryazhimskiy and Plotkin 2008), two basic trends emerged from this analysis. First, only a small percentage of the B. cereus core genes (∼5% in clade 1 and 2) had identifiable non-neutral selection patterns (Supplemental Table 4). This low rate was also seen recently in another Gram-positive pathogen, Clostridium difficile (He et al. 2010). Second, the genes identified were quite different between clades 1 and 2, perhaps reflecting selection on functions responsible for clade-specific niches. Nevertheless, the lists revealed interesting patterns that may offer clues about the ecological specializations within each clade. Amino acids metabolism genes under selection in each taxon (ilvC, hisAF, leuD, aroK in clade 1; argH, ilvC in clade 2; argBDJ, cysH in clade 3) may result from adaptation to specific deficiencies in the host environment. Some genes under selection in clade 1 had functions that have been indirectly linked to mammalian pathogenesis; for instance, molybdopterin biosynthesis (moaAD) and the corA magnesium transporter are believed to influence macrophage survival in other bacteria (MacGurn and Cox 2007; Zhu et al. 2009). However, most of the B. anthracis functions identified as necessary to in-host survival from recent molecular studies, such as iron siderophores (Maresso et al. 2006; Zawadzka et al. 2009) and the mntA manganese transporter (Gat et al. 2005), did not exhibit signatures of selection.
Chromosomes of anthrax-causing strains are not exceptional within B. cereus s.l
A shift to a profoundly pathogenic lifestyle within a bacterial lineage may result in a relaxation of selection over most of the genome. As an example, Hershberg et al. (2007) showed that Shigella strains have undergone both gene loss and accumulation of nonsynonymous mutations in their recent evolution within E. coli. Considering its enhanced virulence, B. anthracis or its recent ancestors may have undergone a similar evolutionary transition. Therefore, we asked whether signatures of relaxed selection compared with other members of the B. cereus s.l. species could be discerned in B. anthracis, other pXO1-containing bacteria, or clade 1 in general.
Kuo et al. (2009) identified a trend where pairs of bacterial genomes from species of obligate pathogens had higher dN/dS ratios than free-living or facultative pathogen species. This was attributed to the smaller population size reducing the effect of purifying selection. We performed an analysis using a similar methodology, where the median dN/dS ratio of 1612 clusters of aligned core genes of each pairwise combination of the 58 B. cereus s.l. genomes was calculated. dN/dS ratios of clade 1 and clade 2 showed a relationship with genetic distance (as measured by the number of synonymous substitutions) similar to that previously reported for other bacteria (Supplemental Fig. 7; Rocha et al. 2006; He et al. 2010). Past a threshold of genetic difference, in this case about 20–40,000 synonymous substitutions in the 1612 core genes, the median dN/dS for B. cereus s.l. genomes stabilized in the range of 0.03–0.05 (in line with results calculated for stable species in other studies) (Kuo et al. 2009; Hershberg and Petrov 2010). Closely related genomes showed higher dN/dS ratios, although there was greater variation due to smaller sample sizes. This pattern is probably due to a time-lag for selection to remove slightly deleterious nonsynonymous mutations (Rocha et al. 2006).
A number of genomes outside clades 1 and 2 showed unusually high pairwise dN/dS ratios, even when they were relatively distant from each other (Fig. 6). Five of these genomes showed some evidence of loss of genes from the B. cereus extended core (bce98, bce22, bpmyx, bmyc2, and bmyc3) (Fig. 1). This finding agreed with the study of Kuo et al. (2009) in correlating genome decay with high pairwise dN/dS ratios. We disentangled the contribution of each genome to the pairwise dN/dS value using ANOVA. All the clade 3 strains, with the exceptions of bce9, bce17, bce19, and bce20, significantly raised the dN/dS score above the mean for the species when in pairwise combination with other species. Critically, values for GBAA, BACI, and G9241 all fell within 1 SD of the mean for B. cereus s.l. (Fig. 6). Either the pXO1 plasmids were acquired too recently to leave an imprint on the genome or the demographics of anthrax-causing strains were not macroscopically different from other close relatives.
ANOVA analysis for pairwise inverse dN/dS for each genome. The figure shows the results of an ANOVA analysis performed to compare the mean pairwise dN/dS ratios estimated for each of the genomes. The whiskers of each data point box are the minimum and maximum values, and the solid boxes are the 25%–75% percentile ranges. The color of boxes indicates the clade of the genomes (blue, clade 1; red, clade 2; green, clade 3). (Horizontal line) Overall mean of inverse of all the mean pairwise dN/dS ratios (17.94). (Red circles) The three pXO1-containing strains.
Discussion
In this work we place the B. anthracis genome in the context of B. cereus s.l., paying particular attention to the portioning of the genes constituting the pan-genome. One of the most surprising conclusions from our data is that there appear to be very small numbers of genes (probably dwindling to zero with greater sampling) that are found in most members of either clades 1 and 2 but are not part of the core of the whole species. Although there is evidence for significant homologous recombination and horizontal transfer within B. cereus s.l., (Fig. 5) and the finding that only a relatively small number of genes acquired by HGT seems to have become fixed in one clade (Fig. 4), these events have not obscured the strong phylogenetic signal that points to the ancient divergence of the clades (Figs. 1, 5). This raises the question of the nature of the distinct ecological adaptations within the clades that have led them to persist over long evolutionary time scales. Clades may be defined by enrichment of particular classes of noncore genes, rather than by their absolute presence and absence. These patterns should become more apparent as further genomes are sequenced.
In the context of the species, B. anthracis and the other pXO1-containing strains were not unusual in the extent of genome reduction and accumulation of nonsynonymous mutations suggestive of diminished population size. Both these processes have been suggested to be associated with the adaptation to the pathogenic niche. Surprisingly, gene loss and accumulation of nonsynonymous mutations seemed to be occurring primarily outside of the more familiar B. cereus clade 1 and 2 strains associated with mammalian infections, such as anthrax and B. thuringiensis insect pathogens. These outlying strains also have a higher rate of homologous recombination relative to mutation (Didelot et al. 2009). The only one of these strains with a known association to pathogenicity is the food poisoning strain B. cereus cytotoxis NVH391-98 (bce98), which expresses a diarrheic Nhe hemolytic enterotoxin (Lapidus et al. 2008). It is therefore possible that in this case these patterns of genomic change are not specifically associated with the phenotype of virulence for mammals.
It is well known that genes on the pXO1 plasmid are necessary for anthrax-like pathogenesis; and pXO2-borne capsule genes are necessary for virulence of B. anthracis and maybe B. cereus var. anthracis, but not B. cereus G9241 (Hoffmaster et al. 2004; Klee et al. 2010). Outside the plasmids, there is apparently no gene unique to any of the three strains. Neither has there been homoplasmic loss of patho-adaptive genes. The situation seems to be similar in the insect-killing B. thuringiensis phenotype, which is dependent on the acquisition of the plasmid-encoded δ-endotoxin genes, which has happened through HGT on multiple branches on the phylogeny (Supplemental Data 2). In future studies where many more genomes are available, there may be power to detect any SNPs or other small variants that critically influence virulence of strains that have naturally acquired pXO1. For instance, genetic changes in global regulators and their binding sites may have a profound effect on cellular function (Perez and Groisman 2009), and mutations in outer surface proteins that recognize host epitopes or are targets for immune surveillance and environment-sensing proteins could influence the level of virulence. The contributions to the relatively few genes in each clade under strong selection may also turn out to be critical for pre-adaptation for virulence. However, the main conclusion from this study is that the chromosomes of B. anthracis and the other strains carrying pXO1 and causing anthrax-like disease, CI and G9241, do not stand out from the rest of the species in terms of having undergone major gene loss or shift in selection pressures on the core genome. It is therefore possible that there is no subgroup genetically predisposed to anthrax pathogenesis; instead, any number of B. cereus s.l. or possibly even other Bacillus may be capable of gaining the ability to produce the lethal toxin.
Methods
Bacterial strains
Strains sequenced in this study (Table 1) were propagated at 37°C or 30°C on Brain Heart Infusion media. Isolation of B. cereus strains from soil using Bacillus cereus selective agar followed previously described methods (Read et al. 2010). DNA for genome sequencing was prepared from overnight broth cultures propagated from single colonies streaked on a Brain Heart Infusion agar plate using the Promega Wizard Maxiprep System (Promega).
Genome sequencing and assembly
Genomes were sequenced using the GS-20 and GS-FLX Instruments (454 Life Sciences [Roche]) (Margulies et al. 2005). Libraries for sequencing were prepared from 5 μg of genomic DNA. The sequencing reads for each project were assembled de novo using the Newbler program with default parameters (version 1.1.03.19; 454 Life Sciences (Roche), Inc.). We implemented a thorough quality analysis pipeline to identify putative misassemblies and other low-quality regions (Supplemental Data 3).
Automated annotation
We used the DIYA Perl-based pipeline (Stewart et al. 2009) for automated annotation of contigs generated by WGS data. As the first step of the DIYA pipeline, the contigs generated by Newbler were mapped against the B. anthracis Ames ancestor sequence (Ravel et al. 2009) using the MUMmer alignment tool to create a concatenated ordered “pseudocontig.” When contigs did not map against the references, they were assigned to the end of the pseudocontig in random order. The pseudocontig was used as a template for the programs GLIMMER (Delcher et al. 2007), tRNASCAN-SE (Lowe and Eddy 1997), and RNAmmer (Lagesen et al. 2007) for prediction of open reading frames and RNA genes, respectively. All proteins were searched against the UniRef50 database (Suzek et al. 2007) using BLASTP (Altschul et al. 1997) and against the NCBI Conserved Domain Database using RPSBLAST (Altschul et al. 1997) with an E-value threshold of 10−10 to record matches.
Whole-genome alignment using MAUVE
Bacillus cereus s.l. genomes were aligned using the MAUVE (Darling et al. 2004) algorithm with default settings. The draft contigs were first reordered against Ames ancestor using the Mauve Contig Mover. Three hundred eighty-five LCBs were greater than the 1500 bp set as the minimum. LCBs for each genome were extracted from the output of the program and concatenated.
Clustering protein orthologs
The complete predicted proteome from all genomes annotated in this study, combined with the proteome of the previously published genomes, was searched against itself using BLASTP with default parameters. We removed short, spurious, and nonhomologous hits by setting a bitscore/alignment length filtering threshold of 0.4 and minimum protein length of 30. Predicted proteins passing this filter were clustered into families based on these normalized distances using the Ortho-MCL algorithm (Li et al. 2003) based on the Markov clustering algorithm (Enright et al. 2002).
Core cluster alignments
The program MUSCLE (Edgar 2004) was used with default settings for multiple sequence alignment (MSA) of the protein-coding genes from the clusters defined by Ortho-MCL. The resulting protein alignments were reverse-translated to codon-based nucleotide alignments using PAL2NAL (Suyama et al. 2006), which used the corresponding DNA sequences for positive selection analysis (see below), and another set of protein alignments were filtered by GBLOCKS (Talavera and Castresana 2007) to remove regions that contained gaps or were highly divergent. The following GBLOCKS settings were used: minimum number of sequences for a conserved position, 30; minimum number of sequences for a flank position, 49; maximum number of contiguous nonconserved positions, 8; minimum length of a block, 10; and allowed gap positions, none.
Phylogenetic reconstruction
For the protein-based phylogeny, we used the results of clustering analysis, selecting protein families that were found to have exactly one member in each of the genomes with the length of each protein in the cluster nearly identical. These protein sequences were aligned using MUSCLE (Edgar 2004), and individual gene alignments were concatenated into a string of amino acids for each genome. Uninformative characters were removed from the data set using GBLOCKS (parameters as above) and a phylogeny reconstructed with PHYLIP (Felsenstein 1989) under a neighbor-joining model. To evaluate node support, a majority rule-consensus tree of 1000 bootstrap replicates was computed.
A binary matrix of the presence or absence of each of the 22,975 genes for each genome was created. This matrix was used to create a network phylogeny using the Neighbor-Net algorithm implemented by the SplitsTree software (Kloepper and Huson 2008).
Analysis of positive selection
Genes under positive selection were identified using codeml as implemented in PAML, version 4.4 (Yang 2007). Two types of tests were implemented in PAML to identify genes under positive selection: Test 1 was carried out using the null model M1a (Nearly-neutral) and the alternative model M2a (positive selection). Test 2 was carried out to identify genes under positive selection in specific branches of the B. cereus tree (branch-site test2 described by Zhang et al. 2005). Test 1 identified genes under positive selection in a single or all the branches of a given phylogeny, while test 2 identified genes under positive selection in the whole-genome (species) tree (Fig. 1). Initially, the inferred whole-genome tree was used for all PAML analyses. For all genes that were identified as being under positive selection, Test 1 and test 2 were re-run to check whether the positive selection results obtained using gene-specific trees differed from the whole-genome tree (Fig. 1). For each test, the likelihood of a model that does not allow positive selection (null model) was compared to a model that allows positive selection (alternative model) using a LRT (Zhang et al. 2005). For branch-specific tests (test 2), one degree of freedom was used to calculate P-values, while for the overall test (test 1), two degrees of freedom were used to calculate P-values. Correction for multiple testing was performed using the Benjamini and Hochberg method (Benjamini and Hochberg 1995) implemented in the software Q-value (Storey 2002).
Analysis of homologous recombination
ClonalFrame (Didelot and Falush 2007), version 1.2, was applied to the genomic regions found by MAUVE to be homologous in all 58 genomes. ClonalFrame was run for a total of 20,000 iterations, with the first half discarded to allow the program to converge and the second half recorded every 10 iterations. Four runs were performed independently and in parallel and were found to be highly congruent in term of the phylogenies reconstructed and recombination events detected. The relative effect of recombination and mutation (r/m) in the whole sample and for each clade was calculated by forming the ratio of the number of substitutions introduced by recombination and mutation for the relevant branches of the phylogeny.
Calculation of π and dN/dS
Core gene nucleotide alignments were parsed by custom bioperl scripts that used Perlymorphism libraries (Stajich and Hahn 2005) in order to calculate π and other diversity statistics. dN and dN were calculated from aligned core genes with the Jukes-Cantor correction applied using the BioPerl module Bio::Align::Statistics. These results were cross-verified against values calculated using PAML tools.
Data access
The annotated genome data have been submitted to the NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank) and the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra). The accession numbers are listed in Table 1.
Acknowledgments
We thank Fergus Priest, Margaret Barker, Phyllis Martin, Teddi Shropshire, Ole Andreas Økstad, Anne-Brit Kolstø, and Michel Gohar for sharing strains. We thank Kent Lohman, Patricia Reilly, and members of the 454 Service Center for their help and advice in completing this manuscript. Brian Osborne helped with submission of data to NCBI. Cheryl Timms Strauss edited the manuscript. This work was supported by a contract to 454 Life Sciences, Inc., from the Defense Threat Reduction Agency, and by grant TMTI0068_07_NM_T from the Joint Science and Technology Office for Chemical and Biological Defense (JSTO-CBD), Defense Threat Reduction Agency Initiative, to T.D.R. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the U.S. Department of the Navy, U.S. Department of Defense, or the U.S. Government. Some of the authors are employees of the U.S. Government, and this work was prepared as part of their official duties. Title 17 USC §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 USC §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Footnotes
-
↵13 Corresponding author
E-mail tread{at}emory.edu
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.134437.111.
Freely available online through the Genome Research Open Access option.
- Received November 6, 2011.
- Accepted May 21, 2012.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 3.0 Unported License), as described at http://creativecommons.org/licenses/by-nc/3.0/.