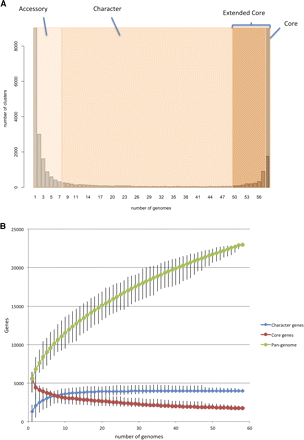
B. cereus pan-genome. (A) Distribution of gene families across B. cereus s.l. genomes The graph of the number of protein clusters present in B. cereus s.l. genomes. Based on the classification of Lapierre and Gogarten (2009), we defined the extended core as genes encoding proteins present in 49 or more genomes. Accessory genes were present in less than six genomes. The class between these extremes defined the character gene set. The core found in every B. cereus s.l. genome comprised 1754 genes (8% of the total gene clusters). There were a further 2148 genes present in the total extended core of 3904 (17% of the total). These genes may be part of the core excluded by the gene-calling software or sequencing errors in one or more WGS genomes, or were lost in nodes of the B. cereus phylogeny undergoing genome reduction (such as the cytotoxic outgroup strain bce98) (Lapidus et al. 2008). These figures for the core and pan-genome size concur with early estimates by Lapidus et al. (2008) and Han et al. (2006). (B) Rarefaction of pan-genome, character, and core genome estimates. The pan-genome and core genome plots (Tettelin et al. 2005, 2008) were based on protein clustering by Ortho-MCL (Methods). The number of gene families present in the pan-genome or core for n number of genomes was calculated based on 100 trials of genomes inputted in random order. Each point of the median size of the set bars represents maximum and minimum values.











