Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells
- 1 Center for Integrative Genomics (CIG), Faculty of Biology and Medicine, University of Lausanne, Lausanne 1015, Switzerland;
- 2 Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland
-
↵3 These authors contributed equally to this work.
Abstract
Our view of the RNA polymerase III (Pol III) transcription machinery in mammalian cells arises mostly from studies of the RN5S (5S) gene, the Ad2 VAI gene, and the RNU6 (U6) gene, as paradigms for genes with type 1, 2, and 3 promoters. Recruitment of Pol III onto these genes requires prior binding of well-characterized transcription factors. Technical limitations in dealing with repeated genomic units, typically found at mammalian Pol III genes, have so far hampered genome-wide studies of the Pol III transcription machinery and transcriptome. We have localized, genome-wide, Pol III and some of its transcription factors. Our results reveal broad usage of the known Pol III transcription machinery and define a minimal Pol III transcriptome in dividing IMR90hTert fibroblasts. This transcriptome consists of some 500 actively transcribed genes including a few dozen candidate novel genes, of which we confirmed nine as Pol III transcription units by additional methods. It does not contain any of the microRNA genes previously described as transcribed by Pol III, but reveals two other microRNA genes, MIR886 (hsa-mir-886) and MIR1975 (RNY5, hY5, hsa-mir-1975), which are genuine Pol III transcription units.
Three main RNA polymerases transcribe the nuclear genome in mammalian cells. RNA polymerase (Pol) I transcribes the large ribosomal RNAs; Pol II the mRNA-coding genes, some small nuclear (snRNA) genes, and regulatory RNA genes such as microRNA genes; and Pol III transcribes a number of noncoding RNA genes whose products are involved in fundamental cellular processes, such as protein synthesis, RNA processing, transcription and chromatin regulation, and in some still poorly defined functions (Schramm and Hernandez 2002; Dieci et al. 2007). Pol III transcription is regulated with cell growth and proliferation, and in response to stress (for review, see White 2005; Goodfellow and White 2007). Deregulation of Pol III is linked to diseases such as Alzheimer's disease, fragile X syndrome, and cancer (for review, see White 2008). Indeed, not only has it been shown that tumor suppressors inhibit Pol III activity, overexpression of a tRNA Met initiator gene is sufficient to cause oncogenic transformation of mouse embryo fibroblasts (Marshall et al. 2008).
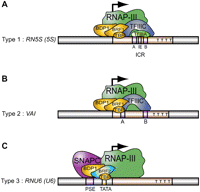
Despite the essential role of Pol III, our knowledge of the Pol III transcription machinery relies largely on biochemical studies performed on essentially three model genes with type 1, 2, and 3 promoters, namely the RN5S (5S), Adenovirus 2 VAI, and mammalian RNU6 (U6) snRNA genes, respectively. As summarized in Figure 1, the internal control region (ICR) of the type 1 5S promoter binds the zinc finger protein TFIIIA. This leads to recruitment of the TFIIIC complex and BRF1–TFIIIB, a factor composed of the TATA box binding protein TBP, the SANT domain protein BDP1, and the TFIIB-related factor BRF1. In contrast, the A and B boxes of the type 2 VAI promoter bind TFIIIC directly, leading to recruitment of BRF1–TFIIIB. In type 3 promoters, the proximal sequence element (PSE) and TATA box that constitute the core region bind, respectively, the snRNA activating protein complex SNAPc and the TBP component of a variant of TFIIIB, BRF2–TFIIIB. BRF2–TFIIIB differs from BRF1–TFIIIB by the replacement of BRF1 with BRF2 (see Schramm and Hernandez 2002). A large scale chromatin immunoprecipitation followed by analysis on a DNA array carrying about 1000 genomic fragments enriched in promoter sequences (ChIP-on-chip) revealed co-occupancy of many Pol III promoters by BRF1, BDP1, and Pol III, suggesting a wide use of these factors in human cells (Denissov et al. 2007). Nevertheless, how broadly this machinery is used in the entire mammalian genome remains to be determined.
Three types of Pol III promoters and corresponding transcription factors. (A–C) Structures of types 1, 2, and 3 promoters, respectively, as well as the factors they recruit.
Another important question is the extent of the Pol III transcriptome. In yeast, ChIP-on-chip studies have defined the yeast Pol III transcriptome as consisting of all tRNA genes and a few additional RNA genes (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). At all these genes, binding of Pol III correlates with gene activity as measured by RNA levels. In Drosophila Schneider line 2 (S2) cells, a recent analysis has identified 359 BRF1 and 354 TRF1 (which replaces TBP in Drosophila TFIIIB) genomic binding sites, corresponding mostly to tRNA genes, as well as to other RNA genes such as the RN7SL1 (7SL) RNA gene and some snoRNA genes (Isogai et al. 2007). In mammalian cells, genome-wide studies have been limited by the lack of a technology to study repeated sequences. Nevertheless, several reports indicate that Pol III may synthesize more RNAs than previously suspected, including microRNAs (Borchert et al. 2006; Ozsolak et al. 2008) and putative new Pol III genes with type 3 promoters (Pagano et al. 2007; see Dieci et al. 2007). Here we have combined the recently developed ChIP-seq method (genome-wide ChIP followed by ultrahigh throughput sequencing of the immunoprecipitated material) with a bioinformatics analysis tailored to deal with repeated sequences to address (1) the generality of the known human Pol III transcription machinery usage and (2) the extent of the Pol III transcriptome, in exponentially growing human IMR90hTert cells.
Results
Localization of POLR3D (RPC4), BDP1, BRF1, and SNAPC2 (SNAP45) in the genome of exponentially growing IMR90hTert cells
Chromatin from IMR90hTert lung fibroblasts, an immortal cell line stably transfected with human telomerase reverse transcriptase, was crosslinked in intact cells, sonicated, and subjected to immunoprecipitation. To identify actively transcribed Pol III transcription units, we used antibodies directed against the POLR3D (RPC4) subunit of Pol III. To identify promoter regions, we used antibodies against BDP1, a hallmark of all three types of promoters, and BRF1, a hallmark of type 1 and 2 promoters (see Fig. 1). For gene-external promoters, we engineered an IMR90hTert cell line expressing the SNAPc subunit SNAPC2 (SNAP45) tagged at its C terminus with the biotin acceptor domain, as well as the biotin ligase BirA (Mito et al. 2005), and performed chromatin affinity purification (ChAP) with streptavidin beads.
The precipitated material was then processed for deep sequencing, whose output is millions of short sequences, referred to as tags (see Supplemental Table S1). Such tags were then mapped onto the genome assembly (UCSC HG18), before being used for peak detection and quantification. In the case of Pol III, tag mapping must take into account large families of highly related genes and pseudogenes. For instance, alignment analyses (BLAT) of RN5S genes reveal 982 related sequences in the human genome. Commonly used methods do not provide a proper solution to this problem in that they (1) use the masked genome assembly from which all repeated sequences are excluded, (2) eliminate tags matching at more than two locations in the genome, and (3) allow up to two sequence mismatches in the alignment of the tags to the genome. Therefore, we devised a custom protocol for analysis of the data, which is described in the Supplemental material. First, we used a partially unmasked genome, which gave us access to repeated sequences. Second, alignment of the tags (35 nucleotides [nt] except for the SNAPC2 experiment, in which we obtained 75-nt tag reads) was performed with the “fetchGWI” software (http://www.isrec.isb-sib.ch/tagger/; Iseli et al. 2007), which allowed us to align tags with multiple matches in the genome. Third, we aligned only tags with a perfect sequence match, which optimized the ability to attribute a tag to the correct genomic location among highly related sequences. Fourth, for peak detection and quantification, we took into account both unique and repeated tags, as described in the Supplemental material.
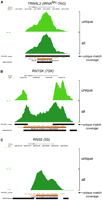
Figure 2 shows UCSC Browser views of three Pol III genes with POLR3D peaks. Figure 2A is an example of a POLR3D peak constituted almost exclusively (99.7%) of tags with unique matches in the genome, which map to a tRNALeu-TAG gene, TRNAU1. The upper track shows the unique tag component and the lower track the combination of unique and repeated tag components. The third track, labeled “unique match coverage,” is represented by a black bar and indicates the DNA regions covered by tags unique within all POLR3D peaks in the genome. In this case, the black bar covers the entire peak region, illustrating that the entire peak width is covered with unique tags (note that the tags are unique within POLR3D peaks and not within the entire genome, as evidenced by the UCSC annotation at the bottom of Figure 2A, showing that part of the peak corresponds to an element repeated in the genome). Consequently, the resulting peaks are identical. Figure 2B is an example with both unique and repeated tags mapping to a RN7SK (7SK) gene. In this case, the unique tag component is represented by two peaks, corresponding to the 5′ and 3′ ends of the RNA-coding sequence, and matched by the unique match coverage track. The combination of unique and repeated tag components results in a continuous peak encompassing the gene. This pattern is consistent with the presence, in the genome, of a number of RN7SK pseudogenes that probably arose by retrotransposition and have, therefore, similar RNA-coding sequences, but divergent flanking sequences. Figure 2C shows an example of a peak composed only of repeated tag reads. It maps to a RN5S gene belonging to a cluster of repeated RN5S genes on chromosome 1; thus, there is no unique tag component in the upper track, and no signal for the unique match coverage track. However, the repeated tag reads form a peak covering the RN5S-coding region. Therefore, in this case, our procedure allowed identification of a gene that would have been missed by a procedure excluding repeated tags. On the other hand, the procedure only allows for approximate quantification, as repeated tags cannot be allocated with absolute certainty to the correct location.
Examples of POLR3D peaks with various proportions of unique and repeated tags. (A–C) UCSC Genome Browser views of the indicated genomic locations. In each case, the upper track labeled “unique” shows the peak obtained with only the unique tag component of the peak shown in the second track, labeled “all,” which contains all tags. The y-axis indicates cumulated tag weights. The track labeled “unique match coverage” shows the regions spanned by tags unique within all POLR3D peaks. Below the unique match coverage, UCSC Browser annotations for RNA genes and repeats from RepeatMasker are shown. The visual peak tracks were generated as described in the legend to Figure 3.
As shown in Supplemental Table S2, the above analysis produced 2169 POLR3D peaks, 1843 BDP1 peaks, and 6353 BRF1 peaks. Peaks found in the input control as well as satellite sequences, simple repeats, and 28S or 18S sequences and related sequences were excluded from our analysis. We chose to avoid the use of stringent filters at this stage, reasoning that we might be able to eliminate noise more specifically by focusing on a combination of peaks containing, at a minimum, a POLR3D peak and either a BDP1 or a BRF1 peak.
POLR3D, BDP1, and BRF1 occupancies mark known Pol III genes
We first examined the location of POLR3D peaks relative to BDP1 and BRF1 peaks, as shown in Table 1. POLR3D peaks were close to both BDP1 and BRF1 peaks in 362 cases, and close to BRF1 or BDP1 peaks in 173 and 174 cases, respectively. To identify genes associated with these peaks, we used the annotations in the various UCSC tables and performed BLAT searches with each known class of Pol III genes to identify any nonannotated genomic sequences resembling them. The resulting genes were divided into four categories as shown in Table 1. The largest category corresponded to tRNA genes, followed by a second category containing some RNA genes (e.g., RPPH1 [RNase P] and RMRP [RNase MRP] genes) and microRNAs, a third category containing the RN5S or RN5S-related sequences, and a fourth category containing Pol II genes (RefSeq genes), Alu repeats, mitochondrial tRNA-derived sequences, and L1 repeats. We started our analysis with the tRNA genes, as this gene family is exceptionally well annotated.
Categories of genes with various combinations of POLR3D (RPC4), BRF1, and BDP1
Location of BDP1, BRF1, and POLR3D peaks on tRNA genes
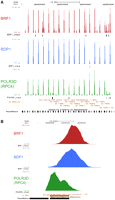
Figure 3A shows a UCSC Genome Browser view of a 600,000-base pair (bp) region of chromosome 6 with a large number of tRNA genes, most of which are associated with BDP1, BRF1, and POLR3D peaks. Figure 3B shows a zoom-in on just two tRNA genes. In both cases, POLR3D peaks cover the whole RNA-coding sequence, whereas BDP1 and BRF1 peaks are located upstream. The peak scores are lower for the gene on the right, indicating lower occupancy. Additionally, we noticed that the BDP1 peaks were shifted further upstream of the transcription start site (TSS) than the BRF1 peaks. Figure 3C shows a plot of peak maxima density relative to TSSs for all tRNA genes displaying BDP1, BRF1, and POLR3D peaks. Most BDP1 peak maxima were at position −27, against position −14 for BRF1. This suggests that within the BRF1–TFIIIB complex, BDP1 contacts DNA slightly upstream of BRF1, consistent with observations in yeast revealing BDP1 crosslinking mostly upstream of BRF1 on the 5′ flanking regions of tRNA genes (for review, see Hernandez 2001). Thus, we can use the chromatin immunoprecipitation with the massively parallel sequencing (ChIP-seq) method to map protein–DNA contacts at high resolution, and we observe that on tRNA genes the BDP1 and BRF1 peak maxima are located within the 5′ flanking region of the genes, largely upstream of the POLR3D peak.
Location of BDP1, BRF1, and POLR3D peaks on tRNA genes. (A) UCSC Genome Browser view of the indicated region of chromosome 6, which contains a number of tRNA genes. The BRF1 and BDP1 visual peak tracks were generated from an overlap profile as described in Zhang et al. (2008). The y-axis indicates cumulated tag weights. For the POLR3D visual track, the same method was used except that the tags were not extended to the actual size of the sequenced fragments to prevent any artificial tag overlaps coming from adjacent peaks reflecting adjacent RNA polymerases. Below each of the BRF1, BDP1, and POLR3D peaks, the unique match coverage is shown, indicating the regions spanned by tags unique within the BRF1, BDP1, and POLR3D peaks, respectively. (B) Zoom-in of a small region of chromosome 6. (C) The density of peak maxima for all tRNA genes displaying BDP1, BRF1, and POLR3D peaks was plotted relative to the TSSs.
Many tRNA genes display low or no BDP1, BRF1, and POLR3D occupancy
The human genome contains 522 predicted tRNA genes and 109 pseudogenes, as annotated by tRNAScan-SE (Lowe and Eddy 1997; Schattner et al. 2005). Of these 631 genes and pseudogenes, 622 have been mapped onto the genome (see http://gtrnadb.ucsc.edu/Hsapi/). Supplemental Table S3 shows these 622 genes and pseudogenes together with their chromosome position, the positions and scores (see below) of the A and B boxes, and the corresponding POLR3D, BDP1, and BRF1 peaks and scores. The genes are sorted by value of the POLR3D peak scores, ranging from 811 to 0.
To evaluate whether POLR3D peak scores reflect transcriptional activity, we measured RNA levels for a few tRNA genes by RT-qPCR. We measured intron-containing tRNA precursors because they are short-lived and, therefore, likely to better reflect ongoing transcriptional activity than mature tRNAs that may have very different stabilities. We analyzed three pairs of precursor tRNAs and as shown in Table 2, we consistently detected more precursor tRNAs for genes with high POLR3D occupancy compared to genes with low or undetectable POLR3D. This suggests that the POLR3D scores reflect, at least to some extent, gene transcriptional activity.
qPCR quantitation of tRNA precursors derived from tRNA genes with different POLR3D (RPC4) peak scores
We then examined the genes in Supplemental Table S3 and divided them into four groups according to the POLR3D scores, as summarized in Table 3. In the first class, containing the genes with POLR3D scores greater than 269, there were no predicted pseudogenes and the large majority of the genes had corresponding BDP1 and BRF1 peaks (93.4% for BDP1 and 97.5% for BRF1). These percentages were still high in the second class of genes with POLR3D scores from 165 to 269, with 76.9% and 90.7% of the genes also containing BDP1 and BRF1 peaks, respectively. This second class contains two predicted pseudogenes; the first (row 122 in Supplemental Table S3) has a BRF1 peak but no BDP1 peak, indeed the number of BDP1 tags in this region is just below the cutoff for peak detection; the second (row 193 in Supplemental Table S3) has clear POLR3D, BDP1, and BRF1 peaks. In both cases, the BDP1 and/or BRF1 peaks are upstream of the POLR3D peak (data not shown). It seems highly likely that these two pseudogenes are in fact expressed, raising the interesting question of the function, if any, of the resulting RNAs. Note also the tRNA SeC(e) gene TRNAU1 (row 138), which displays POLR3D and BDP1 peaks but no BRF1 peak. As discussed further below, this gene displays a SNAPC2 peak and is, therefore, expected to recruit BRF2 rather than BRF1. In the third class, containing genes with POLR3D scores from 3 to 165, 24.4% of the genes also displayed BDP1, and 49.4% also BRF1, peaks. This class contains eight pseudogenes, none of which with BDP1 or BRF1 peaks, suggesting that they are weakly or not transcribed. The fourth class contains 217 genes with no POLR3D peaks, of which 99 are predicted pseudogenes, indicating that the large majority of predicted pseudogenes are not expressed. None of the genes in this class have BDP1 or BRF1 peaks except for one, with the second lowest score BDP1 peak. Thus, at least for tRNA genes, the presence of BDP1 and/or BRF1 is a strong indicator that the gene is also occupied by POLR3D and thus probably transcribed, suggesting that isolated BDP1 or BRF1 peaks elsewhere in the genome do not mark active Pol III transcription units. On the other hand, several tRNA genes displaying POLR3D occupancy show no BDP1 and/or BRF1. The large majority of these genes have, however, low POLR3D scores (<50) suggesting that they are only very weakly transcribed. We note that among tRNA genes, BDP1 and BRF1 scores are, in almost all cases, lower than POLR3D scores. The absence of BDP1 or BRF1 peaks on genes with low POLR3D score is thus consistent with a lower sensitivity of our ChIPs for these two factors. Alternatively, there may exist tRNA genes with no Pol III transcription factors, but with low levels of POLR3D, representing perhaps paused enzymes.
Separation of tRNA genes into four groups according to POLR3D (RPC4) peak scores
Distribution of isoacceptor tRNA genes in the groups with high, medium, low, and no POLR3D occupancy
The annotated human tRNA genes and pseudogenes represent 52 different isoacceptor families (plus two families of suppressor tRNA genes), potentially leading to expression of 52 different isoacceptor tRNAs and two types of suppressors (http://gtrnadb.ucsc.edu/Hsapi/). We examined POLR3D, BDP1, and BRF1 occupancy in each isoacceptor family, and the results are summarized in Supplemental Table S4. The large majority of isoacceptor families are represented in groups 1 or 2, i.e., contain genes with relatively high POLR3D occupancy and accompanying BDP1 and BRF1 peaks, suggesting expression. In contrast, the suppressor tRNA genes and most pseudogenes fall in group 4 and are thus most likely not expressed in dividing IMR90hTert cells. Curiously, in all families containing three or fewer genes and pseudogenes (indicated in purple in Supplemental Table S4), none of the genes harbors convincing POLR3D, BDP1, and BRF1 peaks (the genes always fall in group 4) with two exceptions: the Sec(TCA) genes, with one out of three genes in group 2, and the Ser(ACT) pseudogene, which is the only gene in its family and, as mentioned above, falls in group 2.
tRNA genes with high POLR3D score have slightly different A and B boxes than tRNA genes lacking POLR3D
We wondered whether highly and not expressed tRNA genes might differ in their A and B boxes. Supplemental Table S3 shows the locations and scores of the A and B boxes as determined with the MEME motif search tool (Bailey and Elkan 1994). These scores do not correlate well with POLR3D peak scores (data not shown), suggesting that POLR3D occupancy is dictated by more factors than just the sequence of the A and B boxes, such as, e.g., the spacing of the boxes or elements in the 5′ flanking sequence. Nevertheless, Supplemental Figure S1 shows that the A and B box LOGO outputs of MEME performed on (1) all tRNA genes/pseudogenes in Table S3 (panel A), (2) the tRNA genes with POLR3D scores >400 (panel B), and (3) the tRNA genes (but not the pseudogenes) with no POLR3D (panel C), differ slightly. In the genes with the highest POLR3D occupancy, the A box is extended by one position (−1) at the 5′ end and shortened by one position at the 3′ end, several positions (T1, G3, A7, G11) become fixed, and a number of others have higher information content. Similarly, a number of positions in the B box become fixed (G6, T8, C9, A11, and C14) and others gain in information content. Thus, the A and B box LOGOs for genes with high POLR3D occupancy are the richest in information and may reflect the strongest A and B box components for type 2 promoters.
Other genes with POLR3D, BDP1, and BRF1 peaks: RN5S genes
As shown in Table 1, the second category of genes with combinations of POLR3D, BDP1, and BRF1 peaks corresponded to RN5S genes. We found 982 5S or 5S-related sequences in the genome assembly, either annotated or identified by BLAT searches. However, as shown in Table S5, only 20 5S sequences harbored POLR3D, BDP1, and BRF1 peaks (rows 1–17, 20–22), and another two had POLR3D and BRF1 peaks (rows 18, 19). Seventeen of these, on chromosome 1, correspond to the previously described major 5S cluster on 1q42.11–q42.13 (Steffensen et al. 1974; Sorensen et al. 1991; Stults et al. 2008). Figure 4A shows a UCSC Genome Browser view of this cluster. Each annotated RN5S gene, as well as six additional RN5S sequences identified by BLAT located directly upstream, display BDP1, BRF1, and POLR3D peaks, confirming that, as shown previously in vitro, the 5S promoter recruits BRF1 rather than BRF2 (Wang and Roeder 1995). Note that although these peaks display an average occupancy quasi-identical due to their high sequence identity, some contain point mutations, which are reflected in different peak scores. In contrast to the RN5S genes, two sequences annotated 5S RNA-like, located on either side of the cluster, do not harbor POLR3D, BDP1, or BRF1 peaks. Figure 4B shows an enlargement of one of the RN5S genes. Like for tRNA genes, the BDP1 and BRF1 peak maxima are located upstream of the POLR3D peak, although in this case the BDP1 and BRF1 peak maxima are nearly coincident. This may reflect either a different BRF1 or BDP1 placement on type 1 promoters as compared to type 2 promoters, or differences in crosslinking of these proteins due to the presence of TFIIIA, which might shield or expose BRF1 and BDP1 protein surfaces.
Location of BDP1, BRF1, and POLR3D peaks on RN5S genes. (A) UCSC Genome Browser view of the 5S cluster on chromosome 1. (B) Zoom-in on one of the RN5S gene in the cluster. The visual peak files and the markings are as in the legend of Figure 3.
Of the remaining five RN5S sequences in Supplemental Table S5, two (rows 18 and 19) may be very weakly expressed; they have low POLR3D and BRF1 peaks and lack a BDP1 peak, but the BRF1 peak is clearly located upstream of the TSS (data not shown). In contrast, the other three (rows 20–22) display POLR3D, BDP1, and BRF1 peaks, but these all have maxima inside the RN5S RNA-coding sequence. As these three RN5S RNA-like sequences are very divergent from the true RN5S genes outside of the RNA-coding region, peaks reflecting BDP1 and BRF1 binding should contain unique tags corresponding to the divergent 5′ flanking region. Thus, we suspect that these peaks represent noise generated by our SISSRs (site identification from short sequence reads) analysis.
Other RNA genes and microRNAs
The third category of genes with combinations of POLR3D and BDP1, POLR3D and BRF1, or POLR3D and both BDP1 and BRF1 peaks (see Table 1, category III) contained a number of known RNA coding genes other than tRNA an RN5S genes. We first examined those where the BRF1 peak was present, as genes lacking BRF1 might correspond to genes with type 3 promoters (see Fig. 1). Genes with BRF1 peaks included the vault RNA genes VTRNA1-1 (HVG-1), VTRNA1-2 (HVG-2), and VTRNA1-3 (HVG-3), which encode RNA components of the large cytoplasmic vault particles implicated in multidrug resistance (Kickhoefer et al. 1998), the genes RN7SL1, RN7SL2, and RN7SL3 encoding the 7SL RNA component of the signal recognition particle, a microRNA gene, and the SNAR-A genes, as shown in Supplemental Table S5. These genes are discussed below.
The VTRNA1 (HVG) genes 1–3 differ from the VTRNA1-4 gene by containing not only gene-internal A and B boxes, but also a TATA box and another element located in the 5′ flanking region (van Zon et al. 2001; Kickhoefer et al. 2003). Their occupancy by POLR3D, BDP1, and BRF1 (rows 23–25), and the lack of occupancy on the VTRNA1-4 gene, confirm that the 5′ elements are important for active transcription. Moreover, BRF1 recruitment and the absence of a SNAPC2 peak (see below) show that they are type 2 rather than type 3 genes, despite the TATA box in the 5′ flanking region.
Among a large number of RN7SL-related sequences in the genome, two true genes contain A and B boxes, as well as promoter elements in the 5′ flanking sequence (Ullu and Weiner 1985; Bredow et al. 1990; Kickhoefer et al. 2003; Englert et al. 2004). We found that these two genes (RN7SL1 and RN7SL2) indeed have robust POLR3D, BDP1, and BRF1 peaks (rows 26, 27). In addition, a third gene (RN7SL3, row 28) showed weak POLR3D and BRF1 peaks, but no BDP1 peak, suggesting that it is weakly transcribed.
There are two reports of microRNA genes transcribed by Pol III. Ozsolak et al. (2008) describe a set of 11 microRNAs or microRNA clusters occupied by Pol III or both Pol II and Pol III. We found occupation of one of these, the hsa-mir-565 locus, by BDP1, BRF1, and POLR3D, however, this locus (chr3:45704209–45706624) corresponds, in fact, to a tRNA gene (see http://microrna.sanger.ac.uk/cgi-bin/sequences/mirna_entry.pl?acc=MI0003571; Berezikov et al. 2006). Similarly, we found BDP1, BRF1, and POLR3D on three of the other listed chromosomal locations (chr16:14286104–14288843, chr17:26900553–26902766, and chr7:138675183–138677194), but these also correspond to tRNA genes. None of the other microRNA loci displayed BDP1, BRF1, or POLR3D occupancy in our analysis. Moreover, we noticed that several of these genes contain internal runs of four or even five T residues, which serve as termination signals for Pol III (Bogenhagen and Brown 1981). Thus, these genes are not transcribed by Pol III in exponentially growing IMR90hTert cells, and for some of them at least, it seems unlikely that they will be transcribed by Pol III in other cells (Bogenhagen and Brown 1981). Borchert et al. 2006 identified a microRNA cluster on chromosome 19 (58,860,000 to 58,960,000) with microRNAs synthesized by Pol III from A and B boxes present in adjacent Alu sequences. Additionally, they identified other mir loci with putative Pol III promoters. We did not observe any POLR3D, BDP1, or BRF1 peaks at the locations proposed, except in several cases where the locus contains, in fact, a tRNA gene.
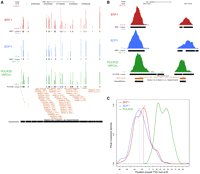
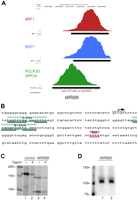
In contrast, we observed clear occupancy of a different microRNA, MIR886 (hsa-mir-886) located on chromosome 5, by BDP1, BRF1, and POLR3D, as illustrated in Figure 5A. This suggests that MIR886 is strongly transcribed by Pol III in IMR90hTert cells, and indeed, as shown in Figure 5B, the DNA sequence corresponding to the POLR3D peak contains A and B boxes, as well as a run of T residues, the termination site for Pol III. We amplified a fragment of genomic DNA containing the sequence occupied by Pol III, as well as flanking sequences, immobilized it on beads, and used it for transcription in vitro. As control, we used a fragment containing the Ad2 VAI gene. As shown in Figure 5C, in vitro transcription of the Ad2 VAI gene fragment gave rise to the expected RNA product, whose synthesis was sensitive to the Pol III-specific inhibitor tagetin, as expected (Fig. 5C, lanes 1, 2). Significantly, with the MIR886 DNA fragment, we obtained an RNA whose size (∼145 nt) and sensitivity to tagetin were consistent with Pol III transcription initiating and terminating as illustrated in Figure 5B (Fig. 5C, lanes 3, 4). A second, shorter fragment (∼100 nt) may correspond to partial processing of the RNA. A Northern blot analysis of RNA extracted from IMR90hTert cells revealed an RNA of ∼100 nt, which corresponds to the length of the predicted pre-miRNA (see http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0005527) (Fig. 5D). Thus, MIR886 is an example of a microRNA genuinely transcribed by Pol III. As described below (Supplemental Table S7), a second example is that of the previously described Pol III RNY5 (hY5) gene, which turns out to overlap with a microRNA gene, MIR1975 (hsa-mir1975).
The MIR886 gene is transcribed by Pol III. (A) UCSC Genome Browser view of the MIR886 microRNA. The visual peak files and the markings are as in the legend of Figure 3. (B) The sequence corresponding to the POLR3D peak region (on the minus strand) is shown with putative A and B boxes indicated (the underlined nucleotides fit the A and B box consensus defined in Supplemental Fig. S1A). The likely transcription start and termination sites, as determined by POLR3D occupancy and sequence, are indicated. (C) The MIR886 gene is transcribed by Pol III in vitro. The genomic fragment carrying the MIR886 locus was immobilized on beads and used as template for in vitro transcription as described in Supplemental material. Tagetin was added in the reactions shown in lanes 2 and 4. The control template was the Ad2 VAI gene. (D) Pre-MIR886 RNA can be detected by Northern blot in IMR90hTert cells. Lanes 1 and 2 show duplicate RNA preparations. The blot was probed with a complementary oligonucleotide corresponding to regions 49–69 and 93–112 of the microRNA gene MIR886.
The small NF90-associated RNA (SNAR) genes encode RNAs associated with NF90 (Parrott and Mathews 2007). There are a number of SNAR genes, 14 SNAR-A, 2 SNAR-B, 5 SNAR-C, and another seven slightly divergent SNAR genes, all but two on chromosome 19. The SNAR-As were shown to be transcribed by Pol III in vitro from an intragenic promoter, and indeed they contain a B box (Parrott and Mathews 2007). We observed relatively low POLR3D and BRF1 peaks on all SNAR-A genes, but not on any of the other SNAR genes. The BRF1 peaks were offset relative to the POLR3D peak and upstream of the TSS, suggesting that these genes are indeed transcribed at a low level in IMR90hTert cells.
Other sequences with POLR3D, BDP1, and BRF1, or POLR3D and BRF1, peaks reveal putative novel genes
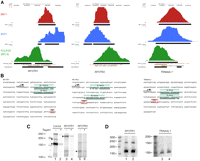
An examination of the sequences in the fourth category of Table 1, labeled “protein coding genes, Alus and other repeats,” revealed that the vast majority of these sequences displayed largely coincident BDP1, BRF1, and POLR3D peaks or combination thereof, which often covered broad sequences. Moreover, the BDP1 and BRF1 peak scores were often much higher than the POLR3D scores, in contrast to what we observe for most bona fide Pol III transcription units. We consider, therefore, that these peaks are probably artifacts and do not correspond to actively transcribed, bona fide Pol III transcription units. There were, however, a few exceptions, which are listed in Supplemental Table S5 (rows 44–50, highlighted in turquoise). Figure 6 and Supplemental Figure S2 show the five most convincing examples. RP3TR1, TRNAAL1, a tRNA-derived sequence, and RP3TR2 contained BDP1, BRF1, and POLR3D peaks (Fig. 6A), whereas POLR3E and MIRb-SINE-MIR contained only BRF1 and POLR3D peaks (Supplemental Fig. S2A). In all cases, the sequence covered by the POLR3D peak contains A and B boxes as well as a run of T residues, as shown in Figures 6B and Supplemental Figure S2B. Figure 6C and Supplemental Figure S2C show transcriptions in vitro performed with genomic fragments containing the RP3TR1, RP3TR2, POLR3E, and MIRb-SINE-MIR sequences, as well as with the control Ad2 VAI gene fragment. In all cases, we obtained RNAs of the expected size and expected tagetin sensitivity. The POLR3E and MIRb-SINE-MIR sequences gave each two RNA products, which may correspond to termination at different runs of T residues (Supplemental Fig. S2C). We tested expression in vivo by Northern blot, as shown in Figure 6D. We could detect expression of TRNAAL1, the tRNA-derived gene (lanes 3, 4), as well as of RP3TR1 (lanes 1, 2). In the latter case, we detected two signals, the slower migrating one consistent with a transcript length corresponding to the POLR3D peak width and the shorter one apparently a spliced form of the first, lacking a short internal region (data not shown). We also detected weak Northern blot RNA signals for the POLR3E and MIRb-SINE-MIR loci, but not for RP3TR2, suggesting that in the latter case the RNA produced is unstable (data not shown). We did not further examine the two remaining putative new Pol III transcription units in Supplemental Table S5 (rows 47, 49). These results reveal active Pol III transcription of a microRNA gene, as well as novel Pol III genes in exponentially growing IMR90h Tert cells.
Examples of three novel Pol III transcription units. (A) UCSC Genome Browser views of three putative new Pol III transcription units (RP3TR1, RP3TR2, and TRNAAL1). The visual peak files and the markings are as in the legend of Figure 3. (B) The sequences corresponding to the POLR3D peak regions (on the minus strand for RP3TR1 and TRNAAL1, and on the plus strand for RP3TR2) are shown. The markings on the sequence are as in the legend of Figure 3. (C) RP3TR1 and RP3TR2 are transcribed by Pol III in vitro. Genomic fragments carrying the RP3TR1 and RP3TR2 loci were immobilized on beads and used as template for in vitro transcription as described in the Supplemental material. Tagetin was added in the reactions shown in lanes 2, 4, and 6. The control template was the Ad2 VAI gene. (D) RP3TR1 and TRNAAL1 RNAs can be detected by Northern blot in IMR90hTert cells. Lanes 1–2 and 3–4 show duplicate RNA preparations. For RP3TR1, the blot was probed with complementary oligonucleotides corresponding to regions 208–237 or 303–328 of the putative gene. For TRNAAL1, the oligonucleotides corresponded to regions 23–46 and 51–71.
SNAPC2 occupancy marks type 3 Pol III genes
As shown in Figure 1, the type 3 RNU6 promoter recruits SNAPc and BRF2–TFIIIB instead of TFIIIC and BRF1–TFIIIB. It seemed thus likely that sequences displaying POLR3D and BDP1 peaks, but no BRF1 peaks, might correspond to genes with type 3 promoters. To help us identify such genes, we performed ChAPs with the IMR90hTert cell line expressing tagged SNAPC2. Here, we describe the SNAPC2 peaks in close proximity to POLR3D peaks. We obtained 35 such peaks, of which 25 were completely overlapping with POLR3D peaks and thus unlikely to mark true Pol III transcription units. We eliminated another two because they lacked a run of at least four T residues at the end of the POLR3D peak. Of the remaining eight, none had close-by BRF1 peaks, as expected for type 3 promoter genes, and six had BDP1 peaks. Supplemental Table S6, rows 1–8, lists the corresponding genomic locations, as well as two additional genomic locations (rows 9, 10) displaying offset BDP1 and POLR3D peaks, a run of T residues at the end of the POLR3D peak, but neither BRF1 nor SNAPC2 peaks. These locations all corresponded to known genes, namely the TRNAU1 (tRNASeC(e) TCA), the RNU6 snRNA, the RNU6ATAC, the RN7SK, the RMRP, the RPPH1, and the RNY1 (hY1) and RNY3 (hY3) genes.
As shown in Supplemental Table S3, only one of three human selenocysteine tRNA genes seemed transcribed (see rows 138, 267, and 496) with POLR3D and BDP1, but no BRF1, peaks. Selenocysteine tRNA genes contain a PSE, a TATA box, and a gene-internal B box, but transcription in vitro does not require the A and B box binding factor TFIIIC (Meissner et al. 1994). The observed binding of SNAPC2, but not BRF1, in vivo confirms that the PSE and TATA box are the functional elements, and thus that the selenocysteine tRNA gene has a functional type 3 promoter.
The human genome contains at least nine, dispersed, full-length RNU6 loci (Domitrovich and Kunkel 2003). Out of these, four (U6-3 to U6-6) lack 5′ promoter elements, and, indeed, U6-4 is not expressed in transfection experiments. In contrast, all others have a DSE, a PSE, and a TATA box and are expressed in transfection experiments, albeit at different levels (Domitrovich and Kunkel 2003; data not shown). We found BDP1, SNAPC2, and POLR3D peaks on RNU6 (U6-8) and RNU6 (U6-9) (Supplemental Table S6, rows 2, 3), and POLR3D and SNAPC2 on RNU6 (U6-2) (Supplemental Table S6, row 4). RNU6 (U6-1), an snRNA gene that has been intensively studied (for review, see Hernandez 2001; Jawdekar and Henry 2008), had surprisingly only a POLR3D peak (peak score 138), and so did RNU6 (U6-7) (peak score 16) (see below; Supplemental Table S7, rows 3, 4). This suggests that three RNU6 snRNA genes are highly transcribed in IMR90hTert cells, with a fourth one (U6-1) transcribed at a lower level and a fifth one (U6-7) transcribed at a very low level.
The RNU6ATAC snRNA is part of the minor U12-type spliceosomal complex involved in splicing of a rare class of introns (the AT–AC introns) (see Lorkovic et al. 2005, references therein). We observed BDP1, POLR3D, and SNAPC2 peaks on a single RNU6ATAC sequence, indicating a type 3 promoter and consistent with the report that there is only one true RNU6ATAC gene in the human genome (Marz et al. 2008). Similarly, we detected SNAPC2, BDP1, and POLR3D on single RN7SK and RMRP sequences, and SNAPC2 and POLR3D on an RPPH1 sequence, consistent with previous reports indicating single true genes with type 3 promoters (Kruger and Benecke 1987; Murphy et al. 1987; Baer et al. 1990; Hannon et al. 1991; Yuan and Reddy 1991).
The Y RNAs associate with the Ro autoantigen to form Ro particles, which have been implicated in UV light resistance and other functions (see Perreault et al. 2007). There are four human Y genes, RNY1 (hY1), RNY3 (hY3), RNY4 (hY4), and RNY5 (hY5) (corresponding to MIR1975, see below), all located on a 50,000-bp region on chromosome 7, and all with type 3 promoters (Wolin and Steitz 1983; Maraia et al. 1994; Maraia et al. 1996). RNY3 and RNY1 code for the longest Y RNAs, whereas RNY4 and RNY5 code for shorter forms. All genes are reported expressed in human cells (Pruijn et al. 1993). We observed BDP1 and POLR3D on the RNY1 and RNY3 genes, and POLR3D peaks only on RNY4 and RNY5 (see below; Supplemental Table S7), suggesting that these last two genes are only weakly expressed.
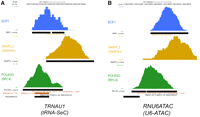
Figure 7 shows the location of the SNAPC2, BDP1, and POLR3D peaks on the TRNAU1 [tRNASeC(e) TCA] and the RNU6ATAC genes. The SNAPC2 peak is well upstream of the TSS, in fact, for all the genes in Supplemental Table S6, the SNAPC2 peak is upstream of the PSE, which is located approximately between positions −50 and −68. Crosslinking experiments suggest that the PSE is bound by the SNAPC4 (SNAP190) and SNAPC3 (SNAP50) subunits of SNAPc. These results suggest that SNAPC2, which associates with the C terminus of SNAPC4 (SNAP190), is located at the “back” of the complex, away from the transcription start site. In contrast, the BDP1 peak is much closer to the TSS.
Location of BDP1, SNAPC2, and POLR3D peaks on the TRNAU1 (tRNA-SeC) and RNU6ATAC genes. UCSC Genome Browser views showing the BDP1, SNAPC2, and POLR3D peaks, as well as the unique tag coverage indicating the regions spanned by tags unique within the BDP1, SNAPC2, and POLR3D peaks, respectively. The POLR3D, BDP1, and SNAPC2 visual peak tracks were generated as described in the legend to Figure 3. (A) TRNAU1 (tRNA-SeC) gene. (B) RNU6 snRNA gene.
POLR3D-only peaks
As discussed above, we chose to focus on combinations of peaks to investigate the extent of the known Pol III transcription machinery in human cells. However, by making this choice we might have discarded bona fide Pol III genes that are weakly expressed or genes whose expression is dependent on unknown transcription factors. To explore these possibilities, we examined the POLR3D peaks lacking neighboring BDP1, BRF1, or SNAPC2 peaks (and not corresponding to tRNA genes). We eliminated those peaks that (1) overlapped sequences containing runs of Ts on either strand, and (2) lacked runs of Ts within a 50-bp window on either side (see Methods). We also eliminated peaks with POLR3D scores smaller than 10 or with shapes never observed on known Pol III transcription units. Supplemental Table S7 lists the remaining peaks. Among them are four known genes already mentioned above (two RNU6 genes, the RNY4 and RNY5 genes). The rest are candidates for novel Pol III genes. To confirm that at least some of these candidate novel genes can in fact serve as Pol III templates, we amplified genomic fragments containing three of them, namely AluJb-SINE-Alu, HuERSP-P1-LTR-ERV1, and MIR-SINE-MIR (rows 13, 18, 69), and used them for transcription in vitro. As shown in Supplemental Figure S3, all three fragments generated RNAs of the expected size and sensitivity to tagetin, consistent with these three loci indeed containing actively transcribed Pol III genes. Moreover, we could detect the expression of AluJb-SINE-Alu in vivo by Northern blot analysis (data not shown). Thus, the three genes from Supplemental Table S7 that we tested are, in fact, actively transcribed, suggesting that several additional candidates listed in Supplemental Table S7 also represent bona fide Pol III transcription units.
Discussion
We have mapped BDP1, BRF1, SNAPC2, and Pol III chromatin occupancy in dividing human IMR90hTert cells by ChIP-seq. To deal with the repeated nature of many Pol III transcription units, we developed an analysis method that takes into account repeated tags. This method can be used to study any genes rich in repeated sequences such as, for example, the Pol II snRNA genes. Rather than eliminating peaks with stringent quantitative filters, as is commonly done in ChIP-seq analyses, we chose to focus on combinations of peaks consisting of POLR3D and at least one of the transcription factors examined, BDP1, BRF1, or SNAPC2, reasoning that such combinations were likely to correspond to bona fide Pol III transcription units. Indeed, examination of factor occupancy on all tRNA sequences revealed that genes with high score POLR3D peaks also harbored BDP1 and BRF1 peaks. As the POLR3D scores diminished, absence of BDP1 or BRF1 peaks became more frequent, with the BDP1 peaks getting lost more quickly than the BRF1 peaks. Importantly, the POLR3D peak scores are likely to reflect transcription efficiency, as in at least the few cases directly examined, genes with high POLR3D peak scores consistently produced more RNA than genes with lower POLR3D occupancy. Thus, the POLR3D ChIPs appear the most sensitive, followed by the BRF1, and then the BDP1, ChIPs. (Because the number of Pol III genes with SNAPC2 peaks is small, it is difficult to assess the relative sensitivity of the SNAPC2 ChAP.)
Examination of tRNA genes revealed that the BDP1 and BRF1 peaks were consistently located upstream of the POLR3D peak, indeed this was true for all known genes with type 1 and 2 promoters and for BDP1 and SNAPC2 on type 3 promoters. Moreover, we found that SNAPC2 was consistently localized upstream of the PSE, whereas previous results have localized SNAPC4 (SNAP190) on the PSE (for review, see Hernandez 2001). This is a remarkable observation, as it indicates that the ChIP-seq method has a resolution high enough to identify different protein–DNA contacts within a complex, and to localize various subunits relative to the DNA. Thus, the ChIP-seq method can be used to dissect, in vivo, protein-DNA contacts between the subunits of a complex on DNA. Here, we used the spatial information as an added criterion to eliminate many peak combinations in which the peaks were completely or largely overlapping. Moreover, we asked that putative Pol III terminators be located at positions compatible with Pol III transcription. As a result, we suspect that there are few false-positives in our analysis.
As we focused our analysis on genomic locations with combinations of POLR3D and at least one of the known Pol III transcription factors, we selected, in effect, for genomic sites with combinations of factors resembling those found on the Pol III model promoters studied so far. It is remarkable that the overwhelming majority of the genomic sites carrying such combinations of factors ended up corresponding to genes known to be transcribed by Pol III. This suggests that the known basal Pol III machinery is used very broadly in actively dividing culture cells. We can now address whether this machinery is also used in differentiated, nondividing cells. Indeed, the basal Pol II machinery seems to undergo major changes in some differentiated cells with replacement of the TFIID complex by another complex containing TAF3 (Deato and Tjian 2007; Deato et al. 2008) (but not TRF3/TBP2, as originally reported [see Gazdag et al. 2009]), raising the possibility of similar major changes in the basal machinery used by other RNA polymerases. We can also address how Pol III occupancy, and thus transcription, varies in different cellular states. We observe, for example, that a large proportion of tRNA genes is poorly or not occupied by POLR3D in dividing IMR90hTert cells. Although the highly POLR3D-occupied tRNA genes have slightly different A and B boxes than tRNA genes with no POLR3D, there is no good correlation between A and B box scores and POLR3D occupancy, suggesting that other factors regulate these genes. Indeed, tissue-specific differences in human tRNA gene expression have been documented (Dittmar et al. 2006). More generally, Pol III transcription in mammalian cells is known to be regulated with cell growth and proliferation by factors such as RB1 (Rb), RBL1 (p107), NOLC1 (p130), TP53, MYC, and MAF1 (for review, see White 2005, 2008; Goodfellow and White 2007). The approach described will allow a global view of the Pol III transcriptome variations with cellular identity and state.
Besides broad use of the basal Pol III transcription machinery, our results reveal that the Pol III transcriptome in actively dividing IMR90hTert cells is only slightly larger than previously suspected, even though we cannot exclude that we are missing weakly transcribed Pol III genes. We were surprised to find that none of the microRNA genes described previously as being transcribed by Pol III harbored BDP1, BRF1, SNAPC2, or POLR3D peaks. Rather, we found that several of the reported microRNA gene genomic positions correspond, in fact, to tRNA genes. Similarly, we checked the genomic locations with putative type 3 promoters (Pagano et al. 2007), but all were negative. On the other hand, we identified a clear Pol III transcribed microRNA, MIR886. Moreover, the RNY5 gene, which has an isolated, but very convincing POLR3D peak, overlaps with MIR1975 (hsa-mir-1975), strongly suggesting that this second microRNA is also produced by Pol III. Eight additional genomic locations, one, TRNAAL1, annotated as tRNA-derived, and the rest with various annotations (Supplemental Tables S5, S7, rows labeled orange in the first column) contain active Pol III transcription units as determined by in vitro transcription and/or Northern blot. Interestingly, of the eight confirmed as actively transcribed genes, three (RP3TR2, AluJb-SINE-Alu, and MIR-SINE-MIR) are located within introns of Pol II transcription units and are transcribed on the opposite strand as the Pol II gene. This raises the possibility that they perform a regulatory function, either at the level of transcription itself, perhaps by reducing the efficiency of Pol II transcription on the other strand, or through an action of their RNA products, which may hybridize to the corresponding pre-mRNA or act at the chromatin level. Another few dozen are likely candidates (Supplemental Table S5, rows 47, 49; Table S7). Thus, there are some 350 tRNA genes (genes in groups 1 and 2 and some in group 3) and ∼130 other genes (genes in Supplemental Tables S5–S7) actively transcribed in dividing IMR90hTert cells. This last group includes a few dozen candidate new Pol III genes, of which we have confirmed nine, notably a microRNA gene, as well as eight genes of unknown function, as novel Pol III transcription units.
Methods
ChIPs
The large scale ChIPs were performed with 5 × 107 million subconfluent IMR90 cells transfected with the human telomerase reverse transcriptase gene (a gift of Greg Hannon, Cold Spring Harbor Laboratory), essentially as described by O'Geen et al. (2006). Minor modifications are detailed in the Supplemental material.
ChAPs
ChAPs were performed with IMR90HTert stable cell line expressing human SNAPC2 fused to the Tobacco etch virus (TEV), the Flag epitope, and the biotin acceptor domain (BAD). The detailed protocol is included the Supplemental material.
Mapping of sequence tags onto the unmasked human genome
The DNA material from the ChIPs was submitted to high-throughput sequencing and sequence tags were generated with the Bustard (Illumina) base caller protocol. The mean size of the fragments sequenced was 115 for POLR3D, 92 for BDP1, 78 for BRF1, and 130 for SNAPC2 ChIPs or ChAPs. The methods used to align tags onto the genome, detect peaks, and quantify peaks are detailed in the Supplemental material.
Acknowledgments
We thank Jérôme Thomas, Sylvain Pradervand, Emmanuel Beaudoing, and Keith Harshman of the Lausanne Genomic Technologies Facility, where the ultrahigh throughput sequencing was performed; Nicolas Guex and Christian Iseli from the Swiss Institute of Bioinformatics (SIB) for advice on fetch GWI usage; Ioannis Xenarios (SIB) for discussion and advice; and Philippe L'Hôte for tissue culture. The computations were performed at the Vital-IT (http://www.vital-it.ch) center for high-performance computing of SIB. This work was funded by the University of Lausanne and SNSF grant 3100A0-109941.
Footnotes
-
↵4 Corresponding author.
E-mail Nouria.Hernandez{at}unil.ch; fax 41-21-692-3925.
-
[Supplemental material is available online at http://www.genome.org. The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE18184.]
-
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101337.109.
- Received October 2, 2009.
- Accepted February 25, 2010.
- Copyright © 2010 by Cold Spring Harbor Laboratory Press
Freely available online through the Genome Research Open Access option.