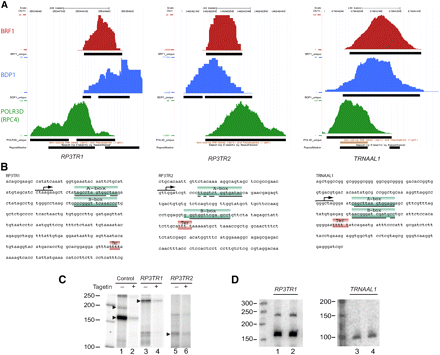
Examples of three novel Pol III transcription units. (A) UCSC Genome Browser views of three putative new Pol III transcription units (RP3TR1, RP3TR2, and TRNAAL1). The visual peak files and the markings are as in the legend of Figure 3. (B) The sequences corresponding to the POLR3D peak regions (on the minus strand for RP3TR1 and TRNAAL1, and on the plus strand for RP3TR2) are shown. The markings on the sequence are as in the legend of Figure 3. (C) RP3TR1 and RP3TR2 are transcribed by Pol III in vitro. Genomic fragments carrying the RP3TR1 and RP3TR2 loci were immobilized on beads and used as template for in vitro transcription as described in the Supplemental material. Tagetin was added in the reactions shown in lanes 2, 4, and 6. The control template was the Ad2 VAI gene. (D) RP3TR1 and TRNAAL1 RNAs can be detected by Northern blot in IMR90hTert cells. Lanes 1–2 and 3–4 show duplicate RNA preparations. For RP3TR1, the blot was probed with complementary oligonucleotides corresponding to regions 208–237 or 303–328 of the putative gene. For TRNAAL1, the oligonucleotides corresponded to regions 23–46 and 51–71.











