Origin of INSL3-mediated testicular descent in therian mammals
- 1 Division of Reproductive Biology, Department of Obstetrics and Gynecology, Stanford University School of Medicine, Stanford, California 94305-5317, USA;
- 2 Chang Gung University School of Medicine, and Department of Obstetrics and Gynecology, Chang Gung Memorial Hospital, Tao-Yuan 333, Taiwan;
- 3 Genome Sequencing Center, Washington University School of Medicine, St. Louis, Missouri 63108, USA
Abstract
Testicular descent is a unique physiological adaptation found in therian mammals allowing optimal spermatogenesis below core body temperature. Recent studies show that INSL3, produced by Leydig cells, and its receptor LGR8 (RXFP2) are essential for mediating the transabdominal phase of testicular descent during early development. However, the origin and genetic basis for this physiological adaptation is not clear. Using syntenic mapping and the functional characterization of contemporary and resurrected relaxin family hormones, we show that derivation of INSL3-mediated testicular descent involved the duplication of an ancestral RLN3-like gene that encodes an indiscriminate ligand for LGR7 (RXFP1) and LGR8. This event was followed by acquisition of the LGR7-selective characteristics by a daughter gene (RLN3) prior to the evolution of the common ancestor of monotremes, marsupials, and placentals. A subsequent mutation of the other daughter gene (INSL3) occurred before the emergence of therian mammals, which then led to the derivation of the reciprocal LGR8-specific characteristics of INSL3. The stepwise evolution of these independent signaling pathways through gene duplication and subsequent divergence is consistent with Darwinian theory of selection and adaptation, and the temporal proximity suggests an association between these genetic events and the concurrent evolution of testicular descent in ancestral therian mammals.
One of the most interesting questions in biology concerns how a novel physiological process evolves in mammals and what are the underlying genetic mechanisms. Recently, it was shown that transabdominal testicular descent is under the direct influence of the INSL3/LGR8 (RXFP2) signaling pathway and is dependent on proper growth of the gubernaculum primordia and the caudal genitor-inguinal ligament (Nef and Parada 1999; Zimmermann et al. 1999; Bogatcheva et al. 2003; Kamat et al. 2004; Wilhelm and Koopman 2006). Curiously, the so-called gubernaculum of Hunter, which extends from the caudal pole of the mesonephric remnants into the inguinal abdominal wall, is common to placentals and marsupials but is absent in monotremata mammals and non-mammalian vertebrates (Griffiths et al. 1993; van der Schoot 1996; Werdelin and Nilsonne 1999; Coveney et al. 2002; Mackay et al. 2004; Wilhelm and Koopman 2006). To date, how testicular descent, and the LGR8-specific INSL3 signaling pathway that underlies this therian mammal-specific adaptation, evolved is not clear.
The generation and subsequent fixation of duplicated genes with previously nonexistent characteristics has been shown to be a principal source of new genes and is associated with great leaps in evolution (Long et al. 2003; Chen et al. 2007). By comparative genomic analyses and point mutations, footprints of molecular adaptations through gene duplication and the subsequent divergence of daughter genes have been found in a handful of duplicated enzymes and receptors involving visual reception, oxygen sensing, steroid hormone signaling, and digestive tract RNA degradation (Yokoyama 2002; Zhang et al. 2002; Long et al. 2003; Thornton 2004; Bridgham et al. 2006). These studies on the cause-effect relationships of gene duplication and biochemical adaptation have focused on proteins that interact with external environmental signals (oxygen for hemoglobins and photons for rhodopsin) or with substrates and ligands not encoded by a gene (foreign RNA for ribonucleases and steroids for steroid receptors). Thus far, no clear case has been made on the cause-effect relationship between the duplication of a polypeptide ligand-mediated signaling network and the evolution of a mammal-specific physiological process (Taylor and Raes 2004; Hughes 2005).
Recently, we identified and characterized relaxin family peptides (human relaxin II [RLN2], INSL3, and relaxin3 [RLN3]) as the cognate ligands for two orphan G protein-coupled receptors (GPCRs), LGR7 (RXFP1) and LGR8 (Hsu et al. 2002; Kumagai et al. 2002; Sudo et al. 2002). Whereas RLN3 and INSL3 are selective agonists for LGR7 and LGR8, respectively, RLN2 activates both LGR7 and LGR8. In addition to signaling through LGR7, RLN3 was shown to be capable of activating two phylogenetically distant GPCRs, GPCR135 (RXFP3) and GPCR142 (RXFP4), but the physiological importance of these interactions remains to be investigated (Liu et al. 2003a, b). Among these ligand-receptor pairs, RLN2/LGR7 signaling is important for the regulation of a subset of therian mammal-specific reproductive processes including parturition through a softened cervix and the development of nipples in females (Zhao et al. 1999; Feng et al. 2005). Disruption of the relaxin (Rln1) or Lgr7 gene in mice led to defects in mammary gland and nipple development and an impaired parturition process (Zhao et al. 1999; Kamat et al. 2004; Krajnc-Franken et al. 2004). On the other hand, it was shown that expression of RLN3 and LGR7 overlaps in several brain regions and that RLN3/LGR7 signaling could be involved in stress and/or memory regulation (Burazin et al. 2005; Ma et al. 2005; Tanaka et al. 2005). In contrast, INSL3/LGR8 signaling was found to be essential for the mediation of testicular descent. Deletion of the Insl3 or Lgr8 gene prevents gubernaculum growth and causes bilateral cryptorchidism in male mice (Nef and Parada 1999; Zimmermann et al. 1999; Bogatcheva et al. 2003), whereas overexpression of INSL3 results in ovary descent to the base of the abdominal cavity due to formation of male-like gubernaculum structures (Adham et al. 2002). These data established that the physiological role of INSL3 is mediated exclusively by LGR8 and that the development of this specific signaling pathway is critical to the evolution of testicular descent in placental mammals. However, it is not clear how INSL3/LGR8-mediated testicular descent evolved in placental and marsupial mammals. Here, based on syntenic mapping and the functional characterization of contemporary and resurrected relaxin family hormones, we identify INSL3 as a signature protein of therian mammals and describe each of the three critical molecular steps associated with the evolution of testicular descent in therian mammals: (1) gene duplication, (2) diversification of the functional components in one of the daughter genes (RLN3), and (3) independent and complementary diversification of the other daughter gene (INSL3).
Results
Relaxin family genes in vertebrates evolved from three independent chromosomal loci in a common ancestor of vertebrates
In humans, there are seven relaxin family genes; however, the number of their orthologs varies greatly among vertebrates (Hsu 1999; Wilkinson et al. 2005b). Several studies that relied on DNA sequence analysis to delineate the evolutionary relationships of relaxin family genes suggested that the diverging inventory of relaxin family genes in different classes of vertebrates could be a result of lineage-specific and genome-wide duplications (Wilkinson et al. 2005b, c). As evidenced by analysis of synonymous (dS) and nonsynonymous (dN) substitution rates, RLN1, RLN2, INSL4, and INSL6 genes, clustered on a 170-kb span of human chromosome 9p24 in tandem (Fig. 1A), were found to be under strong positive selection (Wilkinson et al. 2005b), suggesting that these genes are likely derived from lineage-specific segmental duplications as a result of nonallelic homologous recombinations. In contrast, RLN3 was constrained by purifying selection (Wilkinson et al. 2005b). In addition, these studies indicated that INSL3 shares a close relatedness to RLN1/RLN2/INSL4/INSL6 as compared with RLN3 and INSL5. However, the exact evolutionary path of relaxin family genes in vertebrates remained to be clarified because of heterogeneities in the evolutionary rate as a result of repeated gene duplications and subsequent divergence (Thornton and Kolaczkowski 2005; Wilkinson et al. 2005b, c).
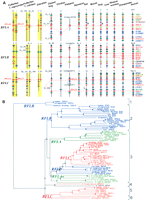
To resolve this conundrum in the phylogenetic reconstruction of relaxin family genes, we analyzed syntenic loci encoding these genes among vertebrates. Mapping of syntenic loci indicated that these genes evolved from three independent loci including (1) INSL5 or relaxin family locus A (RFLA) corresponding to human chromosome 1p31; (2) RLN1/RLN2/INSL4/INSL6 or relaxin family locus B (RFLB) corresponding to human chromosome 9p24; and (3) RLN3/INSL3 or relaxin family locus C (RFLC) corresponding to human chromosome 19p13 in the most recent common ancestor of vertebrates (Fig. 1A). This result is in direct contrast to the inference based on phylogenetic tree analyses that separated tandem-duplicated paralogs at the same locus onto far-flung branches in the tree. For example, an analysis based on 65 relaxin family peptides from 17 species using the maximum likelihood method indicated the presence of six major branches that diverged deep in vertebrate lineage, and orthologous genes derived from each of these three loci were split into at least two distant branches (Fig. 1B; Supplemental Table 1).
Evolution of relaxin family genes in vertebrates. (A) Syntenic mapping of relaxin family gene loci in human (Homo sapiens), chimpanzee (Pan troglodytes), Rhesus monkey (Macaca mulatta), cow (Bos taurus), dog (Canis familiaris), mouse (Mus musculus), rat (Rattus norvegicus), gray short-tailed opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), clawed frog (Xenopus tropicalis), zebrafish (Danio rerio), and two pufferfish (Takifugu rubripes and Tetraodon nigroviridis). The genomes of mammals from human to the egg-laying monotreme platypus encode four to seven paralogous genes. One ortholog of INSL5 on RFLA was identified in all mammals analyzed. In contrast, RFLB contains a single gene in marsupial opossum and monotreme platypus but up to four paralogs in human and chimpanzee (one for each human ortholog RLN1, RLN2, INSL4, and INSL6). On the other hand, orthologs for RLN3 and INSL3 on RFLC were identified in all mammalian species analyzed, except platypus in which the two relaxin family genes on RFLC (RFLCI and RFLCII) exhibited great similarity to RLN3 from other mammals. In contrast, chicken encodes only two relaxin family genes syntenic to RFLA and RFLB in mammals, respectively. Although genes neighboring RFLC could readily be identified on syntenic chicken chromosome 28, no relaxin family gene was detected in this region. On the other hand, the genome of clawed frog encodes four relaxin family genes. Among these paralogs, one each was found to correspond to RFLA and RFLB, respectively. Although the positions of contigs containing the third and fourth genes have not yet been determined, syntenic mapping indicated that they likely represent counterparts of RFLCI and RFLCII in platypus, respectively. The genomes of teleosts including zebrafish and pufferfish encode five copies of relaxin family genes that share close sequence relatedness to mammalian RLN3 (>85% sequence identity at the B chain of mature peptides) (Wilkinson et al. 2005b). Two pairs of these teleost genes were derived from whole-genome duplication (WGD) that occurred before the divergence of teleosts and osteoglossomorphs and correspond to the tetrapod counterpart on RFLA and RFLC, respectively (Jaillon et al. 2004; Crollius and Weissenbach 2005). On the other hand, only one gene syntenic to RFLB was identified, likely representing the remaining member of the WGD-derived ancestral genes on RFLB. Relaxin family genes are indicated by red rectangles, whereas neighboring genes are indicated by diamonds. Orthologous genes in different species are identified by color. Horizontal black boxes on chromosome fragments indicate positions where no overlapping contigs are available in the draft genome sequences. The chromosomal numbers or the genomic contig numbers are indicated at the top of the schematic representation of each genomic fragment. The WGD-derived syntenic chromosomal regions in teleosts are indicated by yellow background. (*) Pseudogene. (B) Phylogenetic analysis of 65 relaxin family peptides from 17 species of vertebrates based on maximum likelihood method. Species analyzed included human (H. sapiens), chimpanzee (P. troglodytes), Rhesus monkey (M. mulatta), rat (R. norvegicus), mouse (M. musculus), rabbit (O. cuniculus), dog (C. familiaris), cow (B. taurus), elephant (L. africana), gray short-tailed opossum (M. domestica), platypus (O. anatinus), chicken (G. gallus), two clawed frogs (X. tropicalis and X. laevis), zebrafish (D. rerio), and two pufferfish (T. rubripes and T. nigroviridis) (accession nos. are provided in Supplemental Table 4). Genes located on chromosome loci syntenic to RFLA, RFLB, and RFLC are indicated by green, blue, and red letters, respectively. The six major branches separated deep in the phylogenetic tree are indicated on the right. The zebrafish sequence without a locus assignment is indicated by black letters.
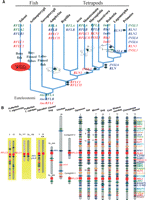
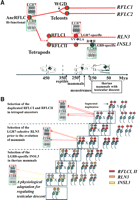
Among these three loci, RFLA remains a single gene locus from teleosts to humans (Figs. 1A, 2A). However, unlike tetrapods that encode a single INSL5 gene, the genomes of teleosts including zebrafish (Danio rerio) and pufferfish (Takifugu rubripes and Tetraodon nigroviridis) encode a pair of co-orthologs on the two syntenic RFLA regions derived from whole-genome duplication (WGD) that occurred before the divergence of teleosts and osteoglossomorphs more than 230–350 million years ago (Mya) (Jaillon et al. 2004; Crollius and Weissenbach 2005). In contrast, RFLB in noneutherian species contains only a single orthologous gene whereas RFLB in hominids encodes up to four paralogs (one for each ortholog of human RLN1, RLN2, INSL4, and INSL6) (Figs. 1A, 2A). Among these genes, RLN1 and INSL4 evolved after the emergence of primates. On the other hand, two paralogous relaxin family genes were identified on RFLC (RFLCI and RFLCII) that are syntenic to human RLN3 and INSL3, respectively, in clawed frog and all mammalian species analyzed (Figs. 1A, 2A). In teleosts, similar to RFLA, one co-ortholog was found on each of the two WGD-derived syntenic RFLC regions of pufferfish. Thus, the most parsimonious evolutionary path for relaxin family genes in vertebrates originated in three separate genes (AncRFLA, AncRFLB, and AncRFLC) in the most recent common ancestor of tetrapods and teleosts >450 Mya (Fig. 2A).
The evolutionary path of relaxin family genes. (A) Syntenic mapping indicated that seven human relaxin family genes evolved from three separate ancestral genes (AncRFLA, AncRFLB, and AncRFLC) in the common ancestor of tetrapods and teleosts. These ancestral genes gave rise to five relaxin family genes in pufferfish (RFLA1, RFLA2, RFLB, RFLC1, and RFLC2) as a result of teleost-specific WGD. One of the WGD-derived RFLB daughter genes (RFLB2) has been lost during teleost evolution. In contrast, a segmental duplication at RFLC led to the generation of RFLCI and RFLCII genes in tetrapods. Whereas RFLCI and RFLCII were lost in avian species, these two genes became RLN3 and INSL3, respectively, in therian mammals. Arrows indicate the putative time of emergence of different relaxin family genes during vertebrate evolution. All genes are color-coded to indicate their origins. Lost genes are indicated by embossed letters. Positions of kidneys and testes in representative vertebrates are indicated by black balls and green balls, respectively. (B) Syntenic mapping of RFLC in vertebrates. The orthologs of human RLN3 and INSL3 on RFLC of different vertebrates are indicated by red rectangles whereas neighboring genes are indicated by diamonds. Orthologous genes in syntenic chromosomal regions of different species are identified by color. The chromosomal numbers or the genomic contig numbers are indicated at the top or the left of the schematic representation of each genomic fragment. WGD-derived syntenic chromosomal regions in teleosts are indicated by yellow background. Genomic fragments derived from segmental duplication of the region neighboring RFLC are indicated by rectangles on the right of the human chromosome 19p13 fragment. (**) The co-orthologous relationship of the zebrafish RFLC2 gene on chromosome 24 to the pufferfish counterparts was determined based on sequence similarity analysis.
INSL3 is a therian mammal-specific signature protein
In addition to revealing the origin of relaxin family genes in vertebrates, syntenic mapping analyses indicated that the neighboring RLN3 (or RFLCI) and INSL3 (or RFLCII) on human chromosome 19p13 were likely derived from a segmental duplication encompassing ∼5–7 megabases of a genomic fragment that included ancestors of EMR, RAB, MYO, PDE4, RAB3, JUN, KLF, CALR, and ILR family genes, and that duplication of the AncRFLC gene predates the radiation of placentals, marsupials, and monotremes but postdates the separation of teleosts and tetrapods (Fig. 2A,B).
Importantly, we found that only therian mammals contain a pair of orthologous INSL3 and RLN3 genes on RFLC (Fig. 2B). The two RFLC genes in platypus and clawed frog (RFLCI and RFLCII) encode peptides with a greater similarity to RLN3 from therian mammals (Fig. 2; Supplemental Table 1). These findings indicate that INSL3 emerged from an RLN3-like ancestor gene on RFLC. Because monotremata mammals are similar to reptiles and other non-mammalian tetrapods in having testes that remain close to the caudal pole of the kidneys throughout life (Fig. 2A), but without a gubernaculum structure or testicular descent (van der Schoot 1996), testicular descent likely emerged in therian mammals in parallel with evolution of the therian mammal-specific INSL3 following separation with monotremata mammals.
Given the finding that RLN3 and INSL3 preferentially interact with LGR7 and LGR8, respectively, it is not obvious how the differential interaction between these two pairs of ligands and receptors could have evolved after duplication of the AncRFLC gene. This enigma is complicated by the asymmetric divergence in their sequences; RLN3 is constrained by strong purifying selection and retains the ancestral sequence features found in species of basal taxonomy, whereas INSL3, similar to RLN1/RLN2/INSL4/INSL6, constitutes many radical substitutions (Wilkinson et al. 2005c). Because LGR7 and LGR8 originated before the evolution of vertebrates, and highly conserved orthologous LGR7 and LGR8 are found in all classes of vertebrates (>75% amino acid similarity is shared by human and teleost LGR7 and LGR8 orthologs), with the exception of chicken, which lacks an LGR8 and an RFLC gene (Hsu et al. 2000; Wilkinson et al. 2005a; Semyonov et al. 2008) (Fig. 1A), we hypothesized that INSL3 and RLN3 could have evolved through a neofunctionalization process in which each of the duplicated daughter genes on RFLC evolved new features that were derived from the original functional characteristics of the parent gene AncRFLC (Force et al. 1999; Postlethwait et al. 2004).
LGR7-specific RLN3 and LGR8-specific INSL3 evolved from a bifunctional ancestral ligand, and the platypus does not contain an LGR8-specific ligand
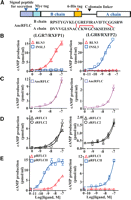
To trace the molecular footprints underlying the evolution of the LGR7-specific RLN3 and LGR8-specific INSL3 signaling pathways, we sought to analyze the functional characteristics of the ancestral peptide on RFLC as well as orthologs from zebrafish and platypus based on LGR7 and LGR8 signaling. From gene resurrection analyses using the maximum likelihood method (Yang et al. 1995; Yang 1997), we generated a series of peptides representing the ancestral RFLC peptides of the common ancestor of vertebrates. Sequence comparison showed that predicted ancestral sequences derived from pooled sequences of non-mammalian species (Supplemental Fig. 1, top) or pools of representative ancestral sequences for each available subclass of vertebrates are in agreement with each other (Supplemental Fig. 1, bottom). The major differences are at the N-terminus of the B chain, which is subjected to alternative processing in different species. Based on these findings, we then generated a recombinant ancestral RFLC peptide (AncRFLC) from the common ancestor of tetrapods and teleosts as well as orthologs from RFLC of zebrafish (zRFLC1 and zRFLC2) and platypus (pRFLCI and pRFLCII) using expression constructs in which the B chain was tagged with a Myc epitope and a 6-histidine epitope (Fig. 3A; Supplemental Fig. 2).
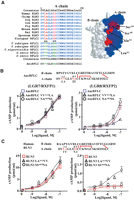
RLN3 and INSL3 evolved from a bifunctional ligand. (A) Schematic presentation of the expression constructs encoding Myc- and 6-histidine-tagged relaxin family peptides linked with a mini-C domain. Mature B and A chain sequences of AncRFLC are shown (bottom). Two inter-chain and one intra-chain disulfide bridges are indicated by red lines. (B–E) LGR7 (RXFP1)- and LGR8 (RXFP2)-activation activities of human RLN3 and INSL3 (B), a resurrected ancestral RFLC peptide (AncRFLC) (C), two zebrafish RFLC peptides (zRFLC1 and zRFLC2) (D), and two platypus RFLC paralogs (pRFLCI and pRFLCII) (E). Mean ± SEM, N = 4.
Unlike human RLN3 and INSL3 that activate only LGR7 and LGR8, respectively (Fig. 3B) (Kumagai et al. 2002; Sudo et al. 2002), analysis of receptor-activation activities showed that the AncRFLC peptide and the zebrafish RFLC1 and RFLC2 peptides are capable of robustly activating both LGR7 and LGR8 from human (Fig. 3C,D; see Supplemental Fig. 2 for Western blotting analyses; see Supplemental Table 2 for pEC50). In contrast, the platypus RFLCI peptide, which corresponds to and clusters with RLN3 of therian mammals in a single branch (Fig. 1A,B), exhibited LGR7-specific characteristics, whereas the RFLCII peptide of platypus, which is syntenic to INSL3, retains ancestral features and activates both LGR7 and LGR8 (Fig. 3E). Therefore, unlike the selective RLN3 and INSL3 genes in humans, the ancestral RFLC gene encoded a bifunctional peptide capable of interacting with both LGR7 and LGR8, and the platypus does not contain an LGR8-specific ligand.
Point mutation of a critical residue in the receptor-binding B chain converts the resurrected AncRFLC ligand from a bifunctional hormone to an LGR8-specific ligand
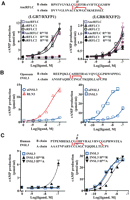
To understand the mechanistic basis for functional divergence of the two bifunctional daughter genes on RFLC of ancestral tetrapods, we sought to identify radical substitutions that are responsible for the functional characteristics of the newly emerged INSL3. Because the B chain of relaxin family peptides represents the major functional domain for interaction with LGR7 and LGR8 (Bullesbach and Schwabe 1991; Rosengren et al. 2006b), we focused the analyses on alternative residues on the B chains of INSL3 and RLN3, which share less than 40% amino acid identity in the mature regions. Comparison of primary sequences and tertiary structures showed that five residues on the B chain (RB12, EB13, FB20, TB21, and SB25) constitute radical substitutions, and three of them (RB12, FB20, and SB25) are on the molecular surface (Fig. 4A). To pinpoint the substitution(s) responsible for the LGR8-specific characteristics of INSL3, we first analyzed mutants of human RLN3 carrying one of each of the three radical substitutions found on the INSL3 surface. Functional analyses showed that substitution of FB20 and SB25 with residues found in corresponding positions in INSL3 had a negligible effect on RLN3 bioactivity (Fig. 4B; Supplemental Table 2). Similar to the wild-type peptide, RLN3 FB20R and RLN3 SB25P mutants function as selective ligands and stimulate cAMP production in cells expressing LGR7 but not LGR8 (Fig. 4B). In contrast, the Arg to His substitution in the RLN3 RB12H mutant abolished the LGR7-activation activity of RLN3 (Fig. 4B, left), and surprisingly, this substitution allowed the RLN3 RB12H mutant to stimulate cAMP production in LGR8-expressing cells (Fig. 4B, right). Thus, the RB12 to HB12 substitution effectively converted RLN3 into an analog with receptor specificity identical to that of INSL3. This conversion in receptor specificity is specific, as an alanine substitution at the same position in the RLN3 RB12A mutant ablates the LGR7-activation activity of RLN3 but without any gain of interaction with LGR8 (Fig. 4B). We confirmed this shift in receptor specificity using receptor-binding assays with 125I-labeled tracers. As shown in Figure 4C, RLN3 and INSL3 compete for labeled RLN2 binding to LGR7 and labeled INSL3 binding to LGR8, respectively (see Supplemental Table 3 for pIC50 values). Unlike wild-type RLN3 that selectively competes for labeled RLN2 binding to LGR7, the RLN3 RB12H mutant competes with labeled INSL3 for binding to LGR8 (Fig. 4C, right), but not with labeled RLN2 for binding to LGR7, on transfected cells (Fig. 4C, left).
RB12H substitution in the B chain of RLN3 converts LGR7-specific RLN3 into an LGR8-specific ligand. (A) Identification of radical substitutions in the INSL3 B chain. Five radical substitutions in INSL3 (green letters). Three surface residues chosen for site-directed mutagenesis analyses (asterisks). Spatial positions of these critical residues are shown (right). In the structure model, residues in the B and A chains are indicated by blue and white space balls, respectively. (B) LGR7(RXFP1)- and LGR8 (RXFP2)-activation activity of human RLN3, INSL3, RLN3 RB12H, RLN3 RB12A, RLN3 FB20R, and RLN3 SB25P peptides (mean ± SEM, N = 4). (C) Competitive LGR7- and LGR8-binding analysis of RLN3, INSL3, RLN3 RB12H, and RLN3 RB12A peptides (mean ± SEM, N = 3). (D) Stimulation of cAMP production in cultured rat gubernaculum cells by INSL3 and RLN3 RB12H peptides (mean ± SEM, N = 4). (*) Significantly different from controls (P < 0.01).
In addition, to investigate whether the gain-of-function RLN3 RB12H mutant is capable of activating native receptors in the gubernaculum, we performed primary cultures of gubernaculum cord cells from 7-d-old neonatal rats and tested the effect of the RLN3 RB12H mutant on gubernaculum function (Kumagai et al. 2002). Consistent with studies of recombinant receptors, treatment of gubernaculum cultures with the INSL3 or RLN3 RB12H mutant increased cAMP production dose dependently (Fig. 4D). In contrast, treatment of the RLN3 or the RLN3 RB12A mutant had no effect on cAMP production in the gubernaculum cells.
Taken together, these studies demonstrated that the RB12 to HB12 mutation represents the critical substitution that occurred during the evolution of INSL3 from the AncRFLC gene. In agreement with this hypothesis, we found that introduction of the receptor-specificity-transforming RB12 to HB12 mutation into the AncRFLC and zebrafish RFLC peptides abolished the LGR7-, but not LGR8-, activation activities of these bifunctional ligands (Fig. 5A). Likewise, functional analyses of opossum INSL3, which represents the most diverging INSL3 ortholog in therian mammals but contains the hallmark HB12 in the receptor-binding B chain, showed that it activates only LGR8 (Fig. 5B).
Ablation of LGR7-activation activity of AncRFLC and restoration of lost LGR7-activation activity in human INSL3 by point mutation. (A–C) LGR7 (RXFP1)- and LGR8 (RXFP2)-activation activity of AncRFLC RB12H, zRFLC1 RB12H, and zRFLC2 RB12H peptides (A), opossum INSL3 peptide (oINSL3) (B), and INSL3 HB12R and INSL3 HB12A peptides (C). Mean ± SEM, N = 4.
Restoration of lost LGR7-activation activity in human INSL3 by an HB12 to RB12 mutation
Apart from the inference that LGR8-specific characteristics of INSL3 are attributable to the RB12 to HB12 mutation, these data predicted that a reversal of this critical substitution in the present-day INSL3 gene could restore the lost function of the parent gene. To investigate this hypothesis, we introduced the RB12 residue found in the AncRFLC peptide into human INSL3 (Fig. 5C). As predicted, substitution of the HB12 with an Arg in the INSL3 HB12R mutant led to restoration of the lost LGR7-activation activity (Fig. 5C, left) without changing the LGR8-activation activity (Fig. 5C, right). In contrast, substitution with an alanine in the same position had negligible effects on the receptor-activating characteristics of INSL3 (Fig. 5C).
Given these results, the most parsimonious course for the evolution of LGR8-specific INSL3 in therian mammals is that the ancestral gene on RFLC encoded a bifunctional ligand, and an Arg to His replacement (a result of two nucleotide changes, CGG to CAT; Supplemental Fig. 3) in one of the duplicated daughter genes eliminated part of the bioactivity (LGR7-activation activity) of the parent gene.
Reciprocal loss of LGR8-activation activity in RLN3 occurred prior to the separation of therian and monotremata mammals
Because the two duplicated daughter genes on RFLC encode bifunctional ligands, the evolution of an LGR7-specific RLN3 in present-day therian mammals likely involved independent mutations. Sequence comparison of mammalian RLN3 and RFLC peptides from non-mammalian vertebrates showed that two pairs of residues at the N terminus of the A chain, A3,4 and A8,9, are on the molecule surface and constitute radical substitutions between the mammalian RLN3 and those from basal taxonomy (Fig. 6A). To pinpoint mutations that are responsible for the shift to LGR7-specific characteristics of mammalian RLN3, we generated mutant AncRFLC peptides carrying substitutions at each of these two pairs of residues (AncRFLC VVA3,4LA and AncRFLC NAA8,9SS; Fig. 6B). Analysis of these mutants showed that point mutations of the N-terminal, but not the C-terminal, pair of residues at the A chain converted the bifunctional AncRFLC peptide into an LGR7-selective agonist (Fig. 6B). To demonstrate further the critical role played by this pair of mutations, we studied RLN3 mutants with reversal substitutions at A3,4 (RLN3 LAA3,4VV) and A8,9 (RLN3 SSA8,9NA) positions (Fig. 6C). Contrary to studies of AncRFLC mutants, functional analyses showed the RLN3 SSA8,9NA mutant remains an LGR7-specific ligand, whereas the RLN3 LAA3,4VV mutant exhibits a gain-of-function activity on LGR8 activation. These data are consistent with studies showing the platypus RFLCI peptide, which contains the LGR7-specific “LA pair” hallmark at the A3,4 positions, activates LGR7, but not LGR8 (Figs. 3E, 6A). Therefore, subsequent to the duplication of AncRFLC, one of the daughter genes, RFLCI, acquired the LGR7-specific characteristics as a result of the VVA3,4 to LAA3,4 mutations in the A chain, and this event occurred before the evolution of proto-, meta-, and eutherian mammals.
Pair of critical substitutions at the A chain ablates LGR8-activation activity of AncRFLC. (A) Sequence comparison and identification of radical substitutions in the mammalian RLN3 A chain. Two pairs of radical substitutions in mammalian RLN3 and platypus RFLCI (asterisks). Spatial positions of these critical residues are shown (right). In the structure model, residues belong to the B and A chains are indicated by white and blue space balls, respectively. (B,C) LGR7 (RXFP1)- and LGR8 (RXFP2)-activation activities of AncRFLC, AncRFLC VVA3,4LA, and AncRFLC NAA8,9SS peptides (B), and RLN3 LAA3,4VV and RLN3 SSA8,9NA peptides (C). Mean ± SEM, N = 4.
Discussion
By mapping of the syntenic chromosomal regions and functional analyses of orthologous relaxin family peptides across vertebrate phyla, we recapitulated the evolutionary footprints of a therian mammal-specific signaling pathway that is crucial to the positioning of the testis from the mesonephric position to the scrotum during embryonic and neonatal development in therian mammals. We showed that the process started with a gene duplication event followed by reciprocal loss of part of the parental functions in the two daughter genes at two separate geological times (Fig. 7A). The stepwise evolution of these independent signaling pathways through gene duplication and subsequent divergence is consistent with Darwinian theory of selection and adaptation, and it suggests an association between the radical substitution of a few key residues of a pair of RLN3-like ancestral genes (RFLCI and RFLCII) and the concurrent evolution of testicular descent in the common ancestor of therian mammals.
Schematic representation of the evolution trajectory of RLN3 and INSL3. (A) RLN3 and INSL3 of therian mammals evolved via gene duplication, followed by emergence of an LGR7 (RXFP1)-specific daughter gene (RFLCI) in the common ancestor of mammals, and an LGR8 (RXFP2)-specific daughter gene (RFLCII) in the common ancestor of therian mammals. LGR7- and LGR8-specific ligands are indicated by a red and a green hexagonal symbol, respectively. Bifunctional ligands are indicated by hexagonal symbols with a mix of red and green colors. Positions of kidneys and testes in representative vertebrates are indicated by black balls and green balls, respectively. (B) Our results support the hypothesis that natural selection selects signaling pathways with a high signal-to-noise ratio over nonselective ones following the duplication of a ligand gene. A high degree of fitness associated with select mutations can then lead to reproductive success and the selection of these copies. In our example, the two daughter genes, RLN3 and INSL3, were selected through natural selection in a stepwise “divergent resolution” manner (Taylor et al. 2001). Starting with a segmental duplication at RFLC, the LGR7-selective RLN3 and the LGR8-specific INSL3 were fixed in the population after neofunctionalization but occurred at two disparate geological time points. Red bars on a chromatid represent RFLCI and RFLCII daughter genes generated by a segmental duplication event in an ancestral tetrapod. Orange bars on a chromatid represent RLN3 after the acquisition of the LGR7-selective characteristics. Gold bars on a chromatid represent INSL3 following the R → H mutation. Genotypes selected by evolution are indicated by rectangular boxes. Ligand specificities for LGR7 and LGR8 are indicated by arrows.
These data demonstrate that subsequent to initial duplication each of the two daughter genes on RFLC (RFLCI and RFLCII) lost part of its parental function to increase the signal-to-noise ratio of the LGR7 and LGR8 signaling pathways mediated by them (Fig. 7B). The reciprocal loss of part of the parental function in a temporally separated manner is necessary as, at each point, one of the daughter genes can afford to lose part of its function because the other daughter gene retains it. In addition, the molecular footprints as described here indicated that INSL3 and RLN3 evolved in a manner similar to the “divergent resolution” model that was proposed to illustrate the separation of different copies of a duplicated gene in allopatric populations during sympatric evolution (Lynch and Conery 2000; Taylor et al. 2001). In this scenario, segregation of the intercellular signaling pathways of the two daughter genes in individual organisms is selected (Fig. 7B), instead of one of the two copies of duplicated genes in reproductively separated populations. Thus, restoration of the lost functions in the RLN3 LAA3,4VV and INSL3 HB12R mutant peptides effectively reversed two critical steps of this ancient “divergent resolution” event that occurred more than 140–180 Mya (Woodburne et al. 2003). Whereas it is generally agreed that newly duplicated genes evolve with rapid changes and many changes in the new genes are needed for their fixation, the discrete interactions between a ligand and a receptor may allow new hormonal functions to be selected and retained with only one or a few changes in these molecules (Long et al. 2003; Bridgham et al. 2006). In addition, we envision the evolution of tissue-specific RLN3/LGR7 and INSL3/LGR8 signaling pathways in therian mammals could be accompanied by a concurrent diversification in the cis-regulatory elements of these ligand and receptor genes.
It has been hypothesized that evolution of transabdominal testicular descent in therian mammals was driven by natural selection for increased aerobic activity associated with endothermy and avoided potential impairment of spermatogenesis when the core body temperature reached a threshold of 34–35°C in primitive mammals (Cowles 1965; Bennett and Ruben 1979). As the transition of the RFLCII gene to INSL3 in ancestral therian mammals (140–180 Mya) occurred much later than the evolution of endothermy in the ancestor of mammals (>220 Mya) (Woodburne et al. 2003; Gillooly et al. 2006), genetic events described here could be associated with a high degree of fitness and reproductive success and contributed to the radiation and dominance of therian mammals in the last 140 million years. Whereas it remains unclear how the male reproductive system in avian species adapted to endothermy, the absence of an RFLC peptide in chicken suggests that alternative mechanisms evolved in avian ancestors to allow optimal spermatogenesis at a high body temperature. Therefore, our study emphasizes the importance of integrating anatomical, physiological, molecular, and phylogenetic data in illustrating the origin and mechanistic bases of gene selection and physiological adaptation.
Furthermore, our investigation provides keys to a fundamental question: How could a novel complex physiological trait or a novel cell type have evolved in a metazoan organism? Because cell type-specific signaling that dictates the proliferation and differentiation of a cell represents one of the most important hallmarks in cell type classification, the duplication of a ligand or its cognate cell surface receptor provided just such a rare opportunity for the generation of a novel intercellular signaling pathway without incurring changes in the fundamental architecture of the cellular components. Our data suggest that the evolution of INSL3 together with the selective expression of LGR8 in an ancestral pregubernaculum cell type allows the emergence of an exclusive signaling pathway for regulating testicular positioning in therian mammals. Therefore, by considering the LGR8-specific INSL3 signaling network as an adaptive structure rather than a signaling pathway, our study provides a paradigm for studying how a lineage-specific physiological trait or a new cell type, such as the gubernaculum cell, could have emerged, as well as how duplications of ligand and receptor genes contributed to physiological complexity during the evolution of metazoans.
Methods
Identification of relaxin family genes
We analyzed the genomic DNA of human (Homo sapiens), chimpanzee (Pan troglodytes), Rhesus monkey (Macaca mulatta), rat (Rattus norvegicus), mouse (Mus musculus), rabbit (Oryctolagus cuniculus), dog (Canis familiaris), cow (Bos taurus), pig (Sus scrofa), elephant (Loxodonta africana), gray short-tailed opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), clawed frog (Xenopus tropicalis), zebrafish, (D. rerio), and two pufferfish (T. rubripes and T. nigroviridis) and identified relaxin family genes by a series of reciprocal sequence comparisons using the BLAST server. Because our analyses allowed the detection of the most divergent INSL4 or even the functionally unrelated insulin, we ruled out the possibility that additional relaxin family genes exist but were not detected in the genomes of studied species. The evolutionary relationships of relaxin family genes were determined using the ClustalW program (http://www.ebi.ac.uk/clustalw). The chromosomal localization of relaxin family genes in different species was determined based on syntenic maps from the Genoscope database (http://www.genoscope.cns.fr) and Ensembl’s BioMart data mining tool (http://www.ensembl.org/multi/martview), followed by verification using the UCSC Genome Bioinformatics web server (http://genome.ucsc.edu/cgi-bin/hgBlat).
Resurrection of ancestral RFLC peptides
Analysis of orthologous RFLC genes as a group indicated that, similar to phylogenetic studies of relaxin family genes as a group, the evolution of RFLC genes in tetrapods mimicked “a radiation event compressed in time” that cannot be resolved by phylogenetic analyses (Thornton 2004; Rokas et al. 2005). We therefore predicted the ancestral RFLC gene sequences using (1) pools of orthologs from non-mammalian species that do not encode an INSL3 ortholog on RFLC to avoid heterotachy incurred by the lineage-specific gene duplication event and subsequent divergence (Thornton and Kolaczkowski 2005) and (2) pools of resurrected ancestral or representative peptide sequences for the different subclasses of vertebrates analyzed (Supplemental Fig. 1). Reconstruction was conducted using codeml(aaml) in the PAML program package (http://abacus.gene.ucl.ac.uk/software/paml.html; Yang 1997).
Comparative structure modeling
Comparative protein modeling was performed with the SWISS-MODEL server (http://swissmodel.expasy.org//SWISS-MODEL.html) using experimentally determined structures for human RLN3 as the template (Protein Data Bank 2FHW) (Rosengren et al. 2006a).
Expression of relaxin family peptides
To generate recombinant RLN3 and INSL3 from different species, we subcloned cDNAs by overlapping PCR with gene-specific primers. Complementary DNA sequences encoding the mature B and A chains were appended with a signal peptide for secretion sequence from the prolactin precursor at the N terminus. In addition, the B chain was tagged with an N-terminal Myc epitope and a C-terminal 6-histidine tag. The B and A chains were connected by a mini-C domain linker of eight amino acids (SLSQEDAL) flanked by dibasic convertase cleavage sites and subcloned into the expression vector, pcDNA3.1 Zeo. The tagging with a Myc and a 6-histidine epitope allowed efficient quantification and purification of recombinant peptides without affecting the bioactivity of the relaxin family peptides. For efficient processing and the secretion of mature peptides, cells were routinely cotransfected with the select expression construct and a one-tenth aliquot of a convertase expression vector (Marriott et al. 1992). Four days after transfection, conditioned media were collected and filtered to remove cell debris. Recombinant peptides were purified using nickel affinity chromatography and quantified based on Western blotting analysis using an anti-Myc antibody (Sigma-Aldrich), followed by densitometry analysis of the immunofluorescent signals. For Western blotting analysis, peptides were resolved using 18% SDS-PAGE and analyzed as previously described (Hsu et al. 2002; Kumagai et al. 2002).
Receptor-activation and receptor-binding analyses
Bioactivity of the relaxin family peptides was determined based on stimulation of the adenylate cyclase activity in HEK293T cells expressing recombinant LGR7 or LGR8 as previously described (Hsu et al. 2002; Kumagai et al. 2002). Because orthologous LGR7 and LGR8 from vertebrates share high polypeptide sequence similarity (>85% similarity among mammalian orthologs and >75% similarity between those of teleosts and humans) and chimerization of LGR7 and LGR8 does not affect the signaling characteristics of these receptors (Sudo et al. 2002), we tested the functional characteristics of different peptides using recombinant human receptors.
To determine the binding characteristics of wild-type and mutant peptides, aliquots of HEK293T cells expressing LGR7 or LGR8 were incubated with increasing doses of purified peptides in the presence of 100 pM of 125I-labeled RLN2 or INSL3 tracer (Phoenix Pharmaceuticals) at room temperature for 2 h. After washing, radioactivity bound to the cells was measured using a gamma counter. Total binding was determined in the absence, and nonspecific binding in the presence, of 100 nmol of unlabeled RLN2 or INSL3.
Gubernaculum cell culture
Gubernaculum cells were isolated by modifying an earlier method (Kumagai et al. 2002). Tissues were removed from 1-wk-old rats and dissociated at 37°C in DMEM/F12 with 0.1% collagenase. Cell debris was removed by passage through a sterile filter, and cells were collected by centrifugation. After culture for 48 h in a 5% CO2 incubator at 37°C, cells were washed and treated with or without hormones in quadruplicate. After 12 h of incubation, total cAMP was measured as previously described (Hsu et al. 2002; Kumagai et al. 2002).
Statistical analysis
Receptor-activation and receptor-binding activity curves generated in nonlinear regression analyses (Prism; GraphPad Software) were evaluated relative to the samples without hormone treatment. In all cases, data are reported as the mean ± SEM of assays in triplicate or quadruplicate. Statistically significant responses (P < 0.01) were determined for each stimulated response to the average nonspecific response from controls using ANOVA and Student’s t-test.
Acknowledgments
We thank Caren Spencer, Cynthia Klein, and Rami Rauch for editorial and technical assistance. We thank Drs. Aaron J.W. Hsueh, Anita Payne, and Marco Conti (Department of Obstetrics/Gynecology, Stanford University) for comments on the manuscript. We also thank Drs. Linda Giudice (Department of Obstetrics, Gynecology and Reproductive Sciences, UCSF) and Jonathan S. Berek (Department of Obstetrics/Gynecology, Stanford University) for the encouragement. C.L.C. also thanks Dr. Yung-Kuei Soong (Department of Obstetrics/Gynecology, Chang Gung Memorial Hospital, Taiwan) for the support and encouragement. J.S. is supported by the Bioinformatics Core of the SCCPRR Center at the Stanford University School of Medicine (NIH HD31398). The authors also acknowledge the support of NIH awards (R21 HD47606 and RO1 DK70652 to S.Y.H.) and a March of Dimes research grant (to S.Y.H.).
Footnotes
-
↵4 Corresponding author.
↵4 E-mail teddyhsu{at}stanford.edu; fax (650) 725-7102.
-
[Supplemental material is available online at www.genome.org.]
-
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7119108.
-
- Received September 7, 2007.
- Accepted February 12, 2008.
- Copyright © 2008, Cold Spring Harbor Laboratory Press