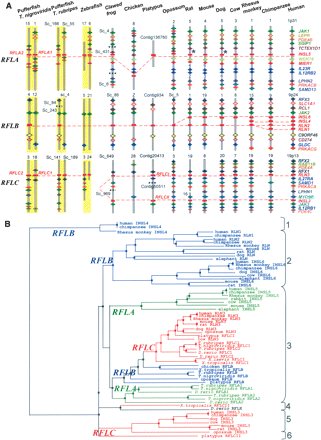
Evolution of relaxin family genes in vertebrates. (A) Syntenic mapping of relaxin family gene loci in human (Homo sapiens), chimpanzee (Pan troglodytes), Rhesus monkey (Macaca mulatta), cow (Bos taurus), dog (Canis familiaris), mouse (Mus musculus), rat (Rattus norvegicus), gray short-tailed opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), clawed frog (Xenopus tropicalis), zebrafish (Danio rerio), and two pufferfish (Takifugu rubripes and Tetraodon nigroviridis). The genomes of mammals from human to the egg-laying monotreme platypus encode four to seven paralogous genes. One ortholog of INSL5 on RFLA was identified in all mammals analyzed. In contrast, RFLB contains a single gene in marsupial opossum and monotreme platypus but up to four paralogs in human and chimpanzee (one for each human ortholog RLN1, RLN2, INSL4, and INSL6). On the other hand, orthologs for RLN3 and INSL3 on RFLC were identified in all mammalian species analyzed, except platypus in which the two relaxin family genes on RFLC (RFLCI and RFLCII) exhibited great similarity to RLN3 from other mammals. In contrast, chicken encodes only two relaxin family genes syntenic to RFLA and RFLB in mammals, respectively. Although genes neighboring RFLC could readily be identified on syntenic chicken chromosome 28, no relaxin family gene was detected in this region. On the other hand, the genome of clawed frog encodes four relaxin family genes. Among these paralogs, one each was found to correspond to RFLA and RFLB, respectively. Although the positions of contigs containing the third and fourth genes have not yet been determined, syntenic mapping indicated that they likely represent counterparts of RFLCI and RFLCII in platypus, respectively. The genomes of teleosts including zebrafish and pufferfish encode five copies of relaxin family genes that share close sequence relatedness to mammalian RLN3 (>85% sequence identity at the B chain of mature peptides) (Wilkinson et al. 2005b). Two pairs of these teleost genes were derived from whole-genome duplication (WGD) that occurred before the divergence of teleosts and osteoglossomorphs and correspond to the tetrapod counterpart on RFLA and RFLC, respectively (Jaillon et al. 2004; Crollius and Weissenbach 2005). On the other hand, only one gene syntenic to RFLB was identified, likely representing the remaining member of the WGD-derived ancestral genes on RFLB. Relaxin family genes are indicated by red rectangles, whereas neighboring genes are indicated by diamonds. Orthologous genes in different species are identified by color. Horizontal black boxes on chromosome fragments indicate positions where no overlapping contigs are available in the draft genome sequences. The chromosomal numbers or the genomic contig numbers are indicated at the top of the schematic representation of each genomic fragment. The WGD-derived syntenic chromosomal regions in teleosts are indicated by yellow background. (*) Pseudogene. (B) Phylogenetic analysis of 65 relaxin family peptides from 17 species of vertebrates based on maximum likelihood method. Species analyzed included human (H. sapiens), chimpanzee (P. troglodytes), Rhesus monkey (M. mulatta), rat (R. norvegicus), mouse (M. musculus), rabbit (O. cuniculus), dog (C. familiaris), cow (B. taurus), elephant (L. africana), gray short-tailed opossum (M. domestica), platypus (O. anatinus), chicken (G. gallus), two clawed frogs (X. tropicalis and X. laevis), zebrafish (D. rerio), and two pufferfish (T. rubripes and T. nigroviridis) (accession nos. are provided in Supplemental Table 4). Genes located on chromosome loci syntenic to RFLA, RFLB, and RFLC are indicated by green, blue, and red letters, respectively. The six major branches separated deep in the phylogenetic tree are indicated on the right. The zebrafish sequence without a locus assignment is indicated by black letters.











