Polymorphisms and the Differential Antiviral Activity of the Chicken Mx Gene
- Jae-Hong Ko1,
- Hee-Kyung Jin1,
- Atsushi Asano1,
- Ayato Takada2,
- Ai Ninomiya2,
- Hiroshi Kida2,
- Hironao Hokiyama3,
- Mutsuo Ohara3,
- Masaoki Tsuzuki4,
- Masahide Nishibori4,
- Makoto Mizutani5, and
- Tomomasa Watanabe1,6
- 1Laboratory of Experimental Animal Science and 2Laboratory of Microbiology, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo 060–0818, Japan; 3Hokkaido Livestock Experiment Station, Shintoku 081–0038, Japan; 4Hiroshima University, Higashihiroshima 739–8528, Japan; 5Nippon Biological Science, Kobuchizawa 408–0041, Japan
Abstract
The nucleotide sequence of chicken Mx cDNA was reported earlier using the White Leghorn breed in Germany, but it showed no enhanced resistance to viruses. In this study, the nucleotide sequences of chicken Mx cDNA were determined in many breeds. A total of 25 nucleotide substitutions, of which 14 were deduced to cause amino acid exchanges, were detected, suggesting that the chicken Mxgene is very polymorphic. Transfected cell clones expressing chickenMx mRNA were established after the Mx cDNA was constructed with an expression vector and introduced into mouse 3T3 cells, and the Mx genes from some breeds were demonstrated to confer positive antiviral responses to influenza virus and vesicular stomatitis virus. On the basis of the comparison among the antiviral activities associated with many Mx variations, a specific amino acid substitution at position 631 (Ser to Asn) was considered to determine the antivirally positive or negative Mx gene. Thus, a single amino acid substitution influences the antiviral activity of Mx in domesticated chickens.
Mx genes are found in a variety of organisms, including yeast (Rothman et al. 1990) and vertebrates ranging from fish to humans (Staeheli et al. 1989; Staeheli 1990; Pavlovic and Staeheli 1991). Mx proteins are antiviral, GTPase enzymes induced by interferon (IFN) (Staeheli 1990; Nakayama et al. 1991, 1992; Samuel 1991; Horisberger 1992; Pitossi et al. 1993). The proteins contain putative tripartite GTP-binding sites as well as a leucine zipper with two elements (Horisberger et al. 1990; Melen et al. 1992; Pitossi et al. 1993). The molecular mechanisms by which the proteins inhibit virus replication appear to be dependent on the subcellular localization. The nuclear Mx1 proteins from the mouse and rat block the replication of influenza virus (Staeheli et al. 1986; Meier et al. 1990). The cytoplasmic Mx2 proteins from the mouse and rat inhibit vesicular stomatitis virus (VSV) but not influenza virus (Meier et al. 1990;Zurcher et al. 1992a). The cytoplasmic human MxA protein confers resistance to influenza virus, VSV, measles virus, and Thogoto virus (Pavlovic et al. 1990; Haller et al. 1993; Schnorr et al. 1993). However, cytoplasmic human MxB and rat Mx3 are without antiviral activity (Meier et al. 1990; Pavlovic et al. 1990). In addition, almost all laboratory mouse strains carry nonfunctional Mx1 andMx2 genes, but feral mouse strains contain functional antiviral Mx1 and Mx2 proteins as shown in our previous studies (Jin et al. 1998; Jin et al. 1999).
In the duck, Mx protein found in the nucleus and cytoplasm showed no enhanced influenza virus resistance (Bazzigher et al. 1993). Furthermore, the chicken Mx protein, which is a predominantly cytoplasmic form, seems to be devoid of antiviral activity (Bernasconi et al. 1995), although the nucleotide sequences of both the Mxstructural gene and its promoter region have been reported (Schumacher et al. 1994; Bernasconi et al. 1995). In this study, we tried to detect polymorphisms of the Mx gene in many breeds of chicken and to find genes antivirally positive to influenza virus and VSV.
RESULTS
Nucleotide and Amino Acid Variations of Chicken Mx
The complete nucleotide sequence of chicken Mx cDNA and the absence of enhanced resistance to viruses were reported earlier (Bernasconi et al. 1995), using the White Leghorn (WLR) breed in Germany. Therefore, we analyzed the nucleotide sequences of chickenMx cDNA from one or two embryos each of many breeds, and compared them with that of the reference breed. We treated fibroblasts from chicken embryos with the IFN inducer-poly (I)/(C) to induceMx mRNA expression. We substantially found the MxmRNA expression only in the culture with poly (I)/(C) and the visualization failed without poly (I)/(C). Numerous nucleotide substitutions at 25 positions were detected in the Mx cDNA, as shown in Table 1. The nucleotide sequence referred to in WLR was virtually unique, because only the sequence from Koshamo (KS) corresponded to it, except for the residue at position 1343 (C to A). The Mx cDNA sequences of other breeds showed 11 to 18 substitutions compared to WLR. A total of 19 independent combinations on the basis of the nucleotide substitutions at 25 residues were observed in the chicken Mx cDNA examined. The Kojidori (KJ), Rhode Island Red (RI), Shamo (SHK, SHL and SHS), Satsumadori (SM), White Leghorn (WLF, WLK and WLO) included nucleotide variations within their Mx cDNA sequences. On the other hand, the same nucleotide sequences were detected in the following groups, Australop (AP) and Fayoumi (GSP); Nagoya (NG), SHS-2, and WLF-1; WLF-2, WLK-2, and WLO-2; and SHK and SHS-1.
Nucleotide Substitutions in Mx cDNA from Various Chicken Breeds
Eleven of the 25 nucleotide substitutions were silent, but the other 14 were deduced to cause amino acid exchanges. Examination of the amino acid exchanges revealed 11 independent combinations of Mx proteins (Table 2). Their relationships are depicted in Figure 1. The amino acid sequence described in WLR was completely consistent with that of the KS breed, but differed in 6 to 12 amino acids from the other breeds. The Mx proteins of AP, Black Minorca (BMC), GSP, NG, SHS-2, WLF-1, and WLK-1 composed a large group showing the same sequence. Furthermore, SM-1, WLF-2, WLK-2, WLO-1, and WLO-2 showed the same sequence of Mx protein and comprised another group. The Mx proteins from the three breeds of Shamo, SHK, SHS-1, and SHL-1 resembled one another and had a distant genetic relationship with the others. SM-2 and Hinaidori (HN) also seemed to have unique sequences.
Amino Acid Substitutions in Mx Protein from Various Chicken Breeds
A phylogenetic tree of the chicken Mx amino acid constructed by using the neighbor-joining (NJ) method.
Permanently Transfected Cell Lines Constitutively Express the Chicken Mx Gene
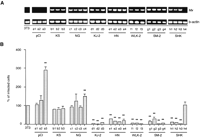
We tried to establish permanently transfected cell lines expressing chicken Mx mRNA at a high level. We chose Mx cDNA from KS, KJ-2, NG, HN, WLK-2, SM-2, and SHK as representatives of the group classified in Figure 1. Their Mx cDNA constructs with plasmid pCI-neo were transfected into the cell line 3T3. 3T3 cells are derived from the BALB/c mouse strain lacking functional Mxgenes and therefore are unable to synthesize endogenous Mx proteins. Individual clones of stably transfected cells containing MxcDNA were tested by reverse transcription (RT)-PCR using primers NE1-F and NE1-R, which amplified a DNA fragment corresponding to nucleotides 124–915 of the Mx cDNA (Fig. 2A). The parental 3T3 cells and those transfected with control pCI-neo had no Mx fragments.
(A) The expression of Mx mRNA in each clone. Total RNA from transfected 3T3 cells was reverse transcribed and PCR amplified using Mx primers (top) and β-actin primers (bottom). 3T3, parental 3T3 cells; pCI (a1–3), control 3T3-pCI-neo; KS (b1–3), 3T3 cells expressing KS MxmRNA; NG (c1–4), 3T3 cells expressing NG Mx mRNA; KJ-2 (d1–3), 3T3 cells expressing KJ-2 Mx mRNA; HN (e1–4), 3T3 cells expressing HN Mx mRNA; WLK-2 (f1–3), 3T3 cells expressing WLK-2 Mx mRNA; SM-2 (g1–4), 3T3 cells expressing SM-2 Mx mRNA; and SHK (h1–3; h4 failed to express theMx mRNA), 3T3 cells expressing SHK Mx mRNA. An aliquot of each PCR product was electrophoresed on a 1.2% agarose gel and visualized with ethidium bromide. (B) The infectivity of VSVΔG*-G in Mx cDNA-transfected cell lines. The infectivity on parental 3T3 cells is expressed as 100%. Shown are mean value ± standard errors of the means (n = 10). Significance levels atP < .01 (**) compared with 3T3 cells are indicated.
Chicken Mx of Some Breeds Confers Antiviral Activity to VSV and Influenza Virus
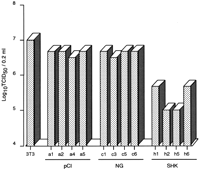
First, using the permanently transfected cell lines, we analyzed the antiviral potential of chicken Mx against VSVΔG*-G infection. Three or four 3T3 cell clones each transfected withMx cDNAs from some chicken breeds were established and examined (Fig. 2). The cell lines expressing Mx from KJ-2, HN, WLK-2, SM-2, and SHK showed positive antiviral responses to VSV infection, with a significantly lower number of infected cells than those of control 3T3 cells, except for one clone (h4) of SHK that showed no amplified Mx fragment in RT-PCR. On the other hand, none of the cell lines expressing Mx from KS and NG could inhibit VSV infection, showing no difference from the control. Furthermore, the same results were observed in an experiment with influenza virus H5N1 in parental 3T3 transfected cells with control pCI-neo, and NG and SHK Mx cDNAs (Fig.3), in which clones a1, a2, c1, c3, h1, and h2 shown in Figure 2 and another two clones from each section were utilized selectively. The cells expressing the SHK Mx showed higher antiviral activity at 48 h postinfection between one and two log units in the 50% tissue culture infective dose (TCID50) against the influenza virus than those with NG Mx and the control.
The infectivity of influenza virus H5N1 in Mx cDNA-transfected 3T3 cell clones. 3T3, parental 3T3 cells; pCI (a1, 2, 4, and 5), 3T3 cells transfected with pCI-neo; NG (c1, 3, 5, and 6), transfected 3T3 cells expressing NG Mx mRNA; and SHK (h1, 2, 5, and 6), transfected 3T3 cells expressing SHK Mx mRNA. Clones a1, a2, c1, c3, h1, and h2 correspond to the designations in Figure 2. Viral titers were determined by the 50% tissue culture infective dose (TCID50) method at 48 h postinfection.
Specific Amino Acid Substitution in 631 (Ser to Asn) Confers to Antiviral Activity
Based on the results shown in Figures 2 and 3, the cells transfected with Mx cDNA from various chicken breeds were divided in antivirally positive and negative groups (Table3). The VSV resistibility and amino acid substitutions at 14 positions were compared between the negative (KS, NG, and WLR) and positive (KJ-2, HN, WLK-2, SM-2, and SHK) groups. Only a specific amino acid substitution at position of 631 (Ser to Asn) emerged to determine a difference between the negative and positive Mx antiviral activity. No correlation with the resistance was observed for the other 13 amino acid substitutions.
Amino Acid Substitution at Position 631 in Mx Protein of Some Chicken Breeds and Its Antiviral Activity against VSVΔG*-G
DISCUSSION
Some mammalian Mx proteins are known to confer a high degree of resistance to certain viruses such as influenza virus and VSV (Staeheli et al. 1986; Meier et al. 1990; Pavlovic et al. 1990; Zurcher et al. 1992a). Chickens are natural hosts to influenza virus (Easterday 1975), and many strains of influenza virus replicate fulminantly in chickens, causing fatal disease. However, it was reported that chicken Mx lacks inhibitory activity toward influenza virus and VSV (Bernasconi et al. 1995). A sensitive chicken Mx gene was demonstrated only in the White Leghorn breed in Germany. In this study, the Mx cDNA sequences were examined in many chicken breeds and were surprisingly shown to be very polymorphic. Furthermore, our results showed that there was a positive antiviral Mx gene in some chicken breeds against influenza virus and VSV. On the other hand, virus-infected 3T3 cells with the Mx cDNA of KS and NG were antivirally negative. Avian cells expressing murine Mx1 protein showed resistance to three strains of influenza A, and three orders of magnitude reduction in influenza virus yield were seen (Garber et al. 1991). The influenza virus H5N1 used in our experiment is the most virulent of subtypes of chicken influenza virus. However, it should be studied more with other subtypes of influenza virus to confirm the antiviral activity of chicken Mx. In 1997, an outbreak of influenza virus in chickens occurred in Hong Kong (Centers for Disease Control and Prevention 1997), and the virus was the likely cause of a human's death (Subbarao et al. 1998). It would be of great value for the poultry industry and also for public health to develop chicken breeds equipped with Mx proteins resistant to RNA viruses.
Our sequence results revealed that an amino acid variation of Asn at position 631 was specific to positive antiviral Mx from KJ-2, HN, WLK-2, SM-2, and SHK. On the other hand, that of Ser was specific to negative Mx from KS, NG, and WLR (Table 3). This single amino acid substitution probably influences the antiviral activity of chicken Mx. Compared with Mxs of other species, chicken and duck Mx proteins in this region near 631 showed high homology of >90%. The duck Mx, which was reported to be nonantiviral (Bazzigher et al. 1993), carries Ser at position 631 like the negative Mx in the chicken. Ser at position 631 was observed in human MxA and MxB, and bovine Mx1. Gly at this position was detected in pig Mx1, equine Mx, and rat Mx1. Furthermore, mouse Mx1 and Mx2 have Arg and His at position 631, respectively. The amino acid of this position is very variable. Asn, present in the chicken Mx showing antiviral activity, was not found in other species. A single amino acid substitution with artificial mutation also was demonstrated in human MxA and mouse Mx1, resulting in different antiviral activities from their normal-type Mx. For example, mutant human MxA (Arg645), which differs from original MxA by a Glu to Arg substitution near the carboxy terminus, is inactive against VSV but active against influenza virus (Zurcher et al. 1992b). Moreover, the cytoplasmic form of mutant Mx1 (Glu614), which differs from native Mx1 by a single substitution in its nuclear transport signal, failed to inhibit the multiplication of influenza virus and VSV (Zurcher et al. 1992c). These observations indicated that even one amino acid substitution in a certain Mx region could change the antiviral specificity.
Mx proteins contain a tripartite consensus motif that typically is found in proteins with GTP-binding and GTP-hydrolyzing activities at the amino terminus (Dever et al. 1987). Mutant human MxA and mouse Mx1 with disrupted GTP-binding motifs were unable to hydrolyze or bind GTP, and failed to confer virus resistance (Pitossi et al. 1993). In addition, all known Mx proteins have leucine repeats, named the leucine zipper, which are expected to be responsible for the oligomerization at the carboxy terminus (Melen et al. 1992). Our current results demonstrated that the GTP-binding domains and leucine repeats were conserved among all polymorphisms of the chicken Mx gene detected in this study. However, Ser to Asn substitution in the region very close to the carboxy terminus was found to determine the antiviral activity of the chicken Mx. In future work, we will try to determine how this substitution influences the antiviral activity of chicken Mx protein at the molecular level, and also whether the virus resistance is determined by the Mx protein in in vivo experiments.
METHODS
Chicken Breed
Fifteen chicken breeds were used in this study. HN, KJ, KS, NG, SHK, SHL, SHS, and SM are Japanese native chickens, and GSP is an Egyptian native chicken. AP, NG, RI, SHK, and WLK were provided by the Hokkaido Livestock Experiment Station, Japan. HN, KJ, KS, SHL, SHS, and SM were obtained from Hiroshima University, Japan. BMC, GSP, WLF, and WLO were provided by the Nippon Institute for Biological Science, Japan.
Cell Culture
Embryonic fibroblasts were established from 11-day chicken embryos (Bernasconi et al. 1995). The fibroblastic cells were seeded in a 10-cm2 tissue culture dish (Falcon Labware; Becton Dickinson), and incubated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco-BRL). Second-passage cultured cells were used for experiments.
Generation of Full-length cDNA
The 5′ and 3′ rapid amplification of cDNA ends (RACE) was used with RT-PCR of mRNAs extracted from cultured fibroblasts after they were treated with the IFN inducer poly (I)/(C) for 7 h (Schumacher et al. 1994), using a Marathon cDNA amplification kit (Clontech) to generate double-stranded cDNA. The double-stranded cDNA was ligated with the Marathon cDNA adaptor and purified on a chromaspin-TE1000 column (Clontech). The 5′- and 3′-RACE was performed by using the double-stranded cDNA as a template with the Mx gene-specific primers NE2-F (5′-CCAGAATGCATCAGAG GTGA-3′, bp 671–690) and NE2-R (5′-TCCTTTCCATGCATT GTCTG-3′, bp 1453–1434), and the adaptor primers AP1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′) and AP2 (5′-ACTCACTATAG GGCTCGAGCGGC-3′). Furthermore, Mx gene-specific primers, FUL-F (5′-ATAGAGCAAGCCAGAA GAACAGCAG-3′, bp 113–137) and FUL-R (5′-GCTTTGACA AGGGTAGGCATATCAG-3′, bp 2432–2408) were generated based on the sequences of progressively amplified 5′- and 3′-RACE to obtain the full-length Mx cDNA. Amplified full-lengthMx cDNA fragments were cloned into pGEM-T Easy vector (Promega).
Sequencing and Characterization of Mx cDNAs
Additional Mx gene-specific primers NE1-F (5′-CA GAAGAACAGCAGAACATG-3′, bp 124–143), NE1-R (5′-CCACAATGATTGTCTCTTTG-3′, bp 915–896), NE3-F (5′-TACACACAAAGCACACACCC-3′, bp 1350–1369), NE3-R (5′-GGATTTTGCAAAGTTCCTCA-3′, bp 2328–2309), NE4-F (5′-CAAAGTTGAAGAAATCGTAT-3′, bp 1541–1560), and NE4-R (5′-AGGACAGTAGAGAGGATGAT-3′, bp 2080–2061) were generated based on the sequences of amplified 5′- and 3′-RACE products and used to get fragments for nucleotide sequencing. The sequences were determined using an ABI Prism dRhodamine terminator cycle sequencing kit and BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer) with an ABI Prism 377 genetic analyzer (Perkin-Elmer). From 8 to 16 clones of full-length cDNA from each breed were used to confirm the sequence, and all of them were sequenced in both directions by use of T7 and sp6 primer. All of the sequences were compared with that of theMx mRNA reported earlier for the White Leghorn breed (Bernasconi et al. 1995), as analyzed with the GENETYX-MAC7.2.0 program.
Phylogenetic Analyses
The phylogenetic analyses were done using the neighbor-joining (NJ) method (Saitou and Nei 1987). Alignment of multiple sequences was done using the CLUSTAL X program (Thompson et al. 1997).
Construction of Chicken Mx Expression Vector and Transfection into 3T3 Cells
The complete coding region of chicken Mx cDNA was cloned into the NotI site of the expression vector pCI-neo (Promega), which contains the human cytomegalovirus immediate-early enhancer/promoter and the neomycin phosphotransferase gene. 3T3 cells (embryonic fibroblasts from BALB/c mouse), purchased from Riken Cell Bank, were grown in DMEM containing 10% fetal bovine serum. After chicken Mx cDNA constructed with the expression vector was transfected into 3T3 cells using Lipofectin (Gibco-BRL), essentially as recommended by the manufacturer, transfected clones were selected in medium containing 500 μg of G418 per milliliter (Pavlovic et al. 1990; Fortunati et al. 1996).
RT-PCR
Total RNA (2 μg) from 3T3 cells was reverse transcribed using 200 U of SUPERSCRIPT II RT (Gibco-BRL) in a total volume of 20 μL. NE1-F and NE1-R were used as primers for the expression of the chickenMx gene in transfected 3T3 cells. As an internal control, mouse β-actin mRNA was used in quantitative RT-PCR experiments. The reaction mixture for RT-PCR contained 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.2 mM deoxynucleoside triphosphate (dNTP), 1 μM of each primer, and 200 U of SUPERSCRIPT II. PCR was performed in a DNA Thermal Cycler (Perkin-Elmer) with Taq polymerase in 1.5 mM MgCl2, 0.2 μM of each primer, and 20 μM of each dNTP, as recommended by the supplier. The cycling profile was comprised of an initial denaturating step for 5 min at 94°C followed by 35 cycles at 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 5 min.
VSV and Influenza Virus Infectious Experiment
Recombinant VSV (VSVΔG*-G) carrying the green fluorescent protein (GFP) gene instead of the G protein gene was kindly provided by Dr. M.A. Whitt (University of Tennessee, Memphis, Tenn.). Infectivity of VSVΔG*-G in 3T3 cell clones was determined by counting the number of GFP-expressing cells in 10–20 microscopic fields (Takada et al. 1997). Influenza virus H5N1 (A/Hong Kong/483/97) taken from a human was used in this study. The viruses were propagated in the allantoic cavities of 11-day-old embryonated chicken eggs for 2 d. TCID50 was determined at 48 h after infection by a cytopathic effect (CPE) assay for infectivity of the influenza virus in 3T3 clones. Viral titers were calculated using the method of Reed and Muench (1938) and described as log units. At least three independent experiments were carried out for each clone.
Statistical Analysis
Data were expressed as means ± standard errors of the means. Statistical significance was evaluated by using Fisher's protected least significant difference test. P values of <.05 were considered statistically significant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵6 Corresponding author.
-
E-MAIL watanabe{at}anim.agr.hokudai.ac.jp; FAX 81–11–706–5106.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.210702. Article published online before print in March 2002.
-
- Received August 15, 2001.
- Accepted January 22, 2002.
- Cold Spring Harbor Laboratory Press