Genomic Dissection of Genotype × Environment Interactions Conferring Adaptation of Cotton to Arid Conditions
- Yehoshua Saranga1,
- Mónica Menz2,
- Chun-Xiao Jiang2,
- Robert J. Wright2,
- Dan Yakir3, and
- Andrew H. Paterson2,4,5
- 1The Hebrew University of Jerusalem, Faculty of Agricultural, Food and Environmental Quality Sciences, Department of Field Crops, Vegetables and Genetics, Rehovot 76100, Israel; 2Department of Soil and Crop Sciences, Texas A&M University, College Station, Texas 77843, USA; 3Weizmann Institute of Science, Department of Environmental Sciences and Energy Research, Rehovot 76100, Israel; 4Applied Genetic Technology Center and the Departments of Crop and Soil Science, Botany, and Genetics, University of Georgia, Athens, Georgia 30602, USA
Abstract
The interaction of genotype with environment is of primary importance in many aspects of genomic research and is a special priority in the study of major crops grown in a wide range of environments. Water deficit, the major factor limiting plant growth and crop productivity worldwide, is expected to increase with the spread of arid lands. In genetically equivalent cotton populations grown under well-watered and water-limited conditions (the latter is responsible for yield reduction of ∼50% relative to well-watered conditions), productivity and quality were shown to be partly accounted for by different quantitative trait loci (QTLs), indicating that adaptation to both arid and favorable conditions can be combined in the same genotype. QTL mapping was also used to test the association between productivity and quality under water deficit with a suite of traits often found to differ between genotypes adapted to arid versus well-watered conditions. In this study, only reduced plant osmotic potential was clearly implicated in improved cotton productivity under arid conditions. Genomic tools and approaches may expedite breeding of genotypes that respond favorably to specific environments, help test roles of additional physiological factors, and guide the isolation of genes that protect crop performance under arid conditions toward improved adaptation of crops to arid cultivation.
Approximately one third of the world's arable land suffers from chronically inadequate supplies of water for agriculture, and in virtually all agricultural regions, yields of rain-fed crops are periodically reduced by drought (Kramer 1980; Boyer 1982). Global climatic trends may accentuate this problem (Le Houerou 1996). Efficient irrigation technologies help to reduce the gap between potential and actual yield; however, because of diminishing fresh water supplies in many regions, genetic improvement of crop productivity under arid conditions (Blum 1988) is necessary as a sustainable and economically viable solution to this problem. The development of drought-tolerant crops has been hindered by low heritability of endpoint measurements such as yield and by lack of knowledge of more precise physiological parameters that reflect genetic potential for improved productivity under water deficit.
Water loss from a plant (transpiration) is an unavoidable consequence of photosynthesis (Cowan 1986), whereby the energy of solar radiation is used for carbon fixation. Carbon enters the leaves of plants as carbon dioxide (CO2), diffusing through epidermal pores (stomata), which also permit water vapor to diffuse out. Although increased transpiration reduces water use efficiency (WUE, the ratio between dry matter [DM] production and water consumption at the whole-plant level or between rates of CO2 fixation and transpiration at the leaf level), it also is a benefit in dissipating excess heat (Cornish et al. 1991; Radin et al. 1994). Water stress and heat stress almost invariably co-occur under arid-region field conditions. The resulting need for a balance between tolerance of heat and drought complicates strategies for manipulating plant water use to improve crop productivity under arid conditions.
A merger of physiology and genetics may improve basic understanding of complex genotype × environment interactions, such as plant response to arid conditions, offering new avenues for crop improvement. Using genetic mapping to dissect the inheritance of different complex traits in the same segregating population can be a powerful means to distinguish common heredity from casual associations between such traits (Paterson et al. 1988). Genetic mapping has been used to identify quantitative trait loci (QTLs) responsible for improved productivity under arid conditions (Agrama and Moussa 1996; Tuinstra et al. 1996; Ribaut et al. 1997). Separately, QTLs have also been reported that confer physiological variations thought to be associated with stress tolerance, such as osmotic adjustment (defined as the active accumulation of solutes in response to water deficit as opposed to passive solute concentration caused by water loss; Morgan 1992; Lilley et al. 1996; Morgan and Tan 1996), WUE (measured either directly or indirectly as a carbon isotope ratio,13C/12C, expressed with a differential notation as δ13C; Martin et al. 1989; Mansur et al. 1993), ash content (Mian et al. 1996, 1998), abscisic acid levels (Quarrie et al. 1994;Tuberosa et al. 1998), stomatal conductance (Ulloa et al. 2000), and various measures of plant water status (Lebreton et al. 1995; Teulat et al. 1998a). However, we are aware of only two prior studies in which productivity and physiological differences were genetically mapped in the same populations, permitting a direct test of the extent to which variation in productivity under arid conditions shares common heredity with specific physiological traits. These two studies found productivity to be unrelated to δ13C (Mansur et al. 1993) or to relative water content (Teulat et al. 1998b).
Valued at ∼$20 billion yearly, cotton originates from wild perennial plants adapted to semi-arid, subtropical environments that experience periodic drought and temperature extremes (Kohel 1974). Modern cotton cultivars are the result of intensive selection to produce large quantities of seed epidermal hairs (“fibers” or “lint”) suitable for mechanical harvesting and processing; this selection has unintentionally narrowed genetic variability for drought tolerance and WUE (Rosenow et al. 1983) within each species. However, considerable variation is found between Gossypium hirsutum (GH) and G. barbadense (GB; Saranga et al. 1998). BothGH and GB are tetraploid (2n = 4x = 52), comprised of A and D subgenomes that appear to have diverged from a common ancestor ∼4 to 11 million years ago and then rejoined in a common nucleus ∼1 to 2 million years ago (Wendel 1989). Virtually all genes in tetraploid cotton are represented by one or more copies in each subgenome, in similar (albeit not identical) chromosomal orders in the two subgenomes (Reinisch et al. 1994) and their diploid ancestors (Brubaker et al. 1999). In this study, two generations of progeny from a cross between the predominant cultivated cotton species, GH and GB, have been grown under well-watered versus water-limited conditions and have been measured for a suite of traits related to plant water status, biomass and economic (fiber) productivity, and economic product (fiber) quality. QTL mapping has been used to test the contributions of selected physiological traits to improved productivity and quality under arid conditions.
RESULTS
Overview of QTLs
Among a total of 161 QTLs detected for the 16 measured traits (listed in Table 1), 102 (63%) showed no significant difference in their effects between well-watered and water-limited conditions. The full details of each QTL will be published under separate cover. Of particular interest to the study of plant adaptation to water-limited environments was the subset of 33 QTLs (described in Table 2) that influenced plant productivity (11 QTLs), physiological traits (five QTLs), or fiber quality (17 QTLs) only in the water-limited treatment, showing no significant differences between GH and GB alleles in the well-watered treatment. Thirteen QTLs (seven, four, and two for productivity, physiology, and quality, respectively) influenced plant performance only under well-watered conditions. Thirteen QTLs (one, three, and nine for productivity, physiology, and quality, respectively) influenced the relative values (ratio of phenotype under water-limited to well-watered conditions), indicating differences in stability of plant performance between the two environments. Herein, we present (Fig. 1) and discuss QTLs associated with productivity and physiological traits under water-limited conditions (including relative values) and their relationship(s) to other QTLs (under either environment).
QTLs in Interspecific Gossypium Populations
Biometrical Parameters of QTLs Affecting Productivity and Physiological Traits of Cotton
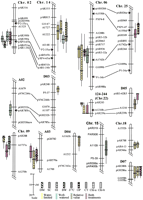
Likelihood intervals for quantitative trait loci (QTLs) implicated in adaptation to arid conditions. The subset of QTLs that were associated with differences between Gossypium hirsutum (GH) andG. barbadense (GB) alleles in productivity (dry matter [DM], seed cotton yield [SC], boll weight [BW], and harvest index [HI]) or physiological traits (carbon isotope ratio [δ13C], osmotic potential [OP], canopy temperature [CT], chlorophyll a content [Chl-a], and chlorophyll b content [Chl-b]) under arid conditions (including relative values) are plotted. In addition, any closely associated QTLs affecting these traits in the well-watered environment or in both treatments are also shown. Traits, environment specificity, and scale in Kosambi centiMorgans are indicated in the legend. Bars and whiskers indicate 1-LOD and 2-LOD QTL likelihood intervals, respectively (Paterson et al. 1988). Nomenclature for chromosomes and linkage groups is as described (Reinisch et al. 1994), except for minor modifications based on new data (J. Rong and A.H. Paterson, unpubl.). Homoeologous chromosome pairs are indicated by lines joining duplicated DNA loci detected by common probes, with the A-subgenome chromosome to the left (except Chr 22–D05, which appears to be an association between two D-subgenome chromosomes; Reinisch et al. 1994). Loci indicated by arrows did not segregate for DNA polymorphisms in this population but are inferred from the primary reference population (Reinisch et al. 1994) to clarify homoeologous alignments. The group designated 124–244 reflects (arbitrary)Mapmaker numbers for the two loci that could be directly mapped in this linkage group – this group is thought to represent Chr 22 based on prior mapping of these loci, but this designation is considered preliminary because it is based on only two loci. To meet space requirements, some chromosomes or linkage groups have been truncated, not showing areas that do not contain relevant QTLs.
QTLs Associated With Plant Productivity
A total of four QTL alleles conferred higher seed cotton yield (SC; closely related to economic yield) under arid conditions (Fig. 1), all of them from GH (chromosome [Chr] 6, Chr 14, Chr 18, LGD07). Three of these QTLs (Chr 14, Chr 18, LGD07) showed no association with any of the measured physiological parameters, but the GHalleles were associated with higher harvest index (the ratio between SC/DM) in the arid environment. One GB QTL allele (Chr 9) conferred higher DM (indicator of total productivity) under arid conditions. Another GH allele (Chr 2) that conferred higher relative DM (indicating a relatively small reduction of productivity in the water-limited environment) was associated with high harvest index and SC under the well-watered treatment and with high boll weight under both environments. We cannot rule out the possibility that this genomic region comprises two or more loci, each accounting for QTLs under a different environment.
QTLs Associated With Physiological Traits
Among three QTL alleles conferring lower osmotic potential (OP), one (Chr 25) under arid conditions and the others under both environments, two (Chr 6 and Chr 25) were also associated with higher SC. These results are further supported by the significant phenotypic correlations between OP and SC (r = −0.28, P<0.001), and OP and DM (r = −0.17, P<0.05) in the water-limited treatment of year 2 (the replicated trial). The likelihood that two of three OP QTLs would be associated with two of five SC QTLs in the cotton genome by chance is ∼0.1% (Larsen and Marx 1985; Lin et al. 1995). Moreover, these two QTLs mapped to corresponding locations on the two different subgenomes of tetraploid cotton (see below), further supporting these findings. This indicates that genetic variation in OP and SC under arid conditions shares a partly common basis. A third QTL allele for reduced OP (LGA05) was associated with increased canopy temperature (CT) under arid conditions.
The genetic control of differences in CT was markedly influenced by water regime. Among four QTLs found to confer genetic differences in CT, one was specific to arid conditions (on linkage group D03) and a second (Chr 6) was specific to relative CT (GH allele conferring higher stability across environments). The GHallele at the Chr 6 CT QTL was associated with higher SC and lower OP. In a genome the size of cotton, this degree of association between CT and yield would occur by chance in ∼7% of cases (Larsen and Marx 1985; Lin et al. 1995; Paterson et al. 1995), offering only tenuous support to the hypothesis that productivity under arid conditions may benefit from dissipation of excess heat (Cornish et al. 1991; Radin et al. 1994). The CT QTL on linkage group D03 showed no influence on productivity.
As was true for CT, the genetic control of chlorophyll content was also markedly influenced by water regime but showed only modest association with productivity. Among three QTLs conferring differences in chlorophyll a, one was specific to the well-watered treatment and one was specific to the relative values (LGA02). Chlorophyll a correlated with DM under the water-limited (r = 0.24, P<0.001) treatment (year 2), but there was no association between QTLs for these traits. Among four QTLs conferring differences in chlorophyll b content, two showed much larger effects in the well-watered treatment. One of these (Chr 14) was associated with SC, in that the GH allele increased both Chlorophyll b under well-watered conditions and SC under dry conditions.
Among 11 QTLs conferring genetic differences in δ13C, three (LGD04, LGD05, Chr 22) were specific to the water-limited environment and one (Chr 15) affected relative δ13C (GH allele conferring higher stability across environments). No δ13C QTLs were associated with differences in biomass (DM) or economic productivity (SC) under water-limited conditions. δ13C correlated with SC (r = 0.19, α = 0.053) in the well-watered treatment of year 1 but not in the water-limited treatment in either year.
Surprisingly, two QTL alleles associated with higher δ13C under arid conditions also coincided with lower chlorophyll b (Chr 22 and LGD05) and chlorophyll a (LGD05). The degree of overlap between these QTLs would be expected to occur by chance in only 0.02% of cases. Because chlorophyll concentration often positively correlates with photosynthetic capacity (Araus et al. 1997), the combination of high δ13C with low chlorophyll concentration possibly indicates low stomatal conductance.
DISCUSSION
The extent to which the inheritance of complex traits differs between well-watered and water-limited conditions reflects the complexity of genotype × environment interactions. The finding (herein; Tuinstra et al. 1997) that partly different sets of loci account for productivity and quality under well-watered versus water-limited conditions (Table 1) indicates that genetic potential for productivity under arid conditions may be improved with little or no penalty under irrigated conditions. At face value, these results seem contradictory to the long-held notion that selection for stress tolerance will generally result in reduced productivity under favorable environments and a decrease in average overall production (Finley and Wilkinson 1963; Rosielle and Hamblin 1981). Our findings might be reconciled with the classical expectation (Finley and Wilkinson 1963;Rosielle and Hamblin 1981) in that simultaneous improvement of productivity (and/or quality) for both arid and irrigated conditions will reduce the expected rate of genetic gain, because of the need to manipulate larger numbers of genes and conduct more extensive field testing (Falconer 1981). These difficulties may be partly ameliorated by efficiencies gained through identification and use of DNA marker–assisted selection (cf. Paterson 1997).
The strategy of crossing two superior genotypes of different species to better exploit the genetic potential for arid-land productivity was borne out by the finding that each of the two species contained different alleles and/or loci conferring adaptation to arid conditions. We crossed GH cv. Siv'on with GB cv. F-177, each of which had the highest WUE among cultivars of their species grown in the test environment in Israel (Saranga et al. 1998). This is contradictory to the prevailing strategy for QTL mapping, which is to choose parental lines with maximal phenotypic divergence (Lander and Botstein 1989). Although elite × elite crosses are typical of traditional plant breeding, interspecific crosses are rarely used in cotton breeding because of numerous barriers to gene flow (Jiang et al 2000). The finding that the GH allele is favorable at some loci and theGB allele at other loci shows that recombination of favorable alleles from each of these species may form novel genotypes that are better adapted to arid conditions than either of the parental species. Marker-assisted selection mitigates many of the problems associated with interspecific crosses (Jiang et al 2000). The genomic exploration of other accessions of these species or other wild tetraploid cottons (G. tomentosum, G. darwinii, and G. mustelinum) may yield still additional valuable alleles. Finally, there also exists considerable variability among “race stocks” (local land races) within G. hirsutum in response to water deficit, which is also well worth further investigation. (Rosenow et al 1983).
The polyploidy of cotton was reflected by two cases in which corresponding homoeologous loci on each of the two different subgenomes appeared to accounts for common sets of traits (Fig. 1). TheGH allele at a QTL on Chr 6 (the A-subgenome) was associated with lower OP, lower CT, and higher SC than was the GB allele in the water-limited environment. At the homoeologous genomic location (on Chr 25), the GB allele conferred both lower OP and higher SC than did the GH allele. A second case of QTLs on homoeologous regions involved GH (Chr 22) and GB(LGD05) alleles that each conferred higher δ13C under the water-limited treatment and lower chlorophyll content under the water-limited or both treatments. The discovery that each of two homoeologous locations account for genetic variation in the same phenotypes indicates that subsequent to polyploid formation in cotton, new functionally significant mutations (alleles) appear to have arisen at each of the two homoeologous loci (or nearby linked loci).
The lack of association of δ13C with productivity under the water-limited environment warrants special attention. δ13C reflects a complex physiological response (Farquhar and Lloyd 1993), specifically an integrated season-long measure of quantitative changes in the relationships between stomatal conductance and photosynthetic capacity, often used as an indicator of WUE in plants (Ehleringer et al. 1993;Condon and Hall 1997). However, high WUE is not necessarily associated with productivity, because plants can modify WUE by different strategies. For example, either increased carbon assimilation rates (at a given stomatal conductance) or reduced stomatal conductance (and transpiration) would enhance WUE, but only the former would increase DM. Water limitation reduced DM and SC to 64% and 68%, respectively, of the control in year 1, and to 47% and 50% in year 2 (based on parental genotypes; data not shown). Our data indicate that under this water deficit, selection for δ13C alone is not expected to improve productivity. The only other study in which both yield and δ13C were mapped (Mansur et al. 1993) supports our finding. We cannot preclude the possibility that more severe water deficits may confer a selective advantage to δ13C: Although such an advantage may improve fitness of wild plants, its economic value in crops may be small or nonexistent.
It is possible that favorable alleles at loci that influence productivity and/or quality only under water-limited conditions may be assembled into genotypes that incorporate adaptations to water-limited conditions but also retain high levels of productivity under well-watered conditions. QTLs that influenced plant performance only under well-watered conditions may be useful for basic research to identify specific metabolic lesions that render some genotypes especially sensitive to water deficit.
Among physiological measures of response to water deficit, our data clearly implicate only OP in adaptation to arid conditions, showing a partly common genetic basis for OP and economic yield under water-limited conditions. In our study, OP determinations were based on leaves sampled at dawn during the boll development period, when irrigation in both treatments had permitted overnight recovery of plant water status. Therefore, differences in OP may have resulted from osmotic adjustment, rather than passive solute concentration caused by water loss (Morgan 1984). A partly common genetic basis adds a new dimension to previously reported phenotypic associations between osmotic adjustment and yield under drought stress (Ludlow at al. 1990;Morgan 1995; El Hafid et al. 1998; Kumar and Singh 1998; Tangpremsri et al. 1995). The importance of osmotic adjustment as an effective mechanism of crop drought resistance is receiving growing recognition (Zhang et al. 1999).
Testing of further traits is needed to account for QTL alleles that have not yet been linked to their physiological basis. For example, three QTLs (Chr 14, Chr 18, LGD07) showed no association with any of the measured physiological parameters, but the GH alleles were associated with higher harvest index in the arid environment, indicating the possible action of mechanisms that favor allocation of photo-assimilates to reproductive organs. The relatively large number of QTLs associated with δ13C may help identify the important physiological traits that contribute to plant stomatal conductance/photosynthetic capacity relationships. Near-isogenic lines are being made for QTLs discovered herein and will offer a powerful tool useful toward identification of the underlying gene(s) by using fine-mapping approaches (Paterson et al. 1990). The availability of cotton bacterial artificial chromosome libraries (C. Abbey, D. Rana, B. Zehr, and A.H. Paterson, pers. comm; D. Peterson, J. Tomkins, R. Wing, and A.H. Paterson, pers. comm.) and established transformation methods for cotton (Bayley et al. 1992), together with the possibility of using comparative approaches (Paterson et al. 1996) to exploit complete sequence data from botanical models such as Arabidopsis, may help to address the complexities of cloning QTLs. Clues as to the physiological roles of the underlying genes may help in designing appropriate probes for parallel high-throughput gene expression studies (Schena et al. 1995; De Risi et al. 1997; Hieter and Boguski 1997; Ruan et al. 1998) and/or mutation searches (Underhill et al. 1997) to identify high-probability candidate genes. The worldwide prevalence (Kramer 1980; Boyer 1982) and possible spread (Le Houerou 1996) of arid lands impel further efforts to dissect the molecular and physiological basis of adaptations to arid conditions in the world's leading crops.
METHODS
Plant Materials
Two field trials were conducted in 1996 and 1997 in Nir-Am, located in the western Negev desert in Israel (31°N, 34°E) each with two irrigation regimes, well watered and water limited. The first experiment consisted of 900 interspecific F2 cotton plants (self-fertilized progenies of a several full-sibling F1hybrid plants from the cross between inbred lines GH cv. Siv'on × GB cv. F-177), grown in 10 main plots (five under each irrigation treatment). About 430 of these plants, which produced sufficient seed for the subsequent experiment, were completely phenotyped and genotyped. Comparison of marker segregation ratios indicated that selection for seed production may have had differential impact on the genome composition of the well-watered versus water-limited environment at some loci; however, in no case was any genotype so rare as to preclude meaningful QTL analysis. The second experiment consisted of 214 F3 families (self-fertilized progenies of the F2, 107 from each treatment to eliminate any possible consequences of differential selection in the F2) selected to represent the entire population, with an emphasis on families with parents that showed extreme values of δ13C. The emphasis on δ13C was in view of the fact that this trait had been associated by many investigators with WUE, and a special priority was to test the role of this trait in productivity and quality of cotton grown under water-limited conditions. Because there were a large number of δ13C QTLs, and these showed only minor overlap with those affecting other traits, this still resulted in a near-random sampling of the genome. A split-plot design was used with irrigation in main plots and with three replicates of five plants per F3 family as subplots. Mean values of the three replicates were used for data analysis. In both experiments, plants were sown in 1.92-m–spaced rows, at a density of four plants per meter. Water was applied twice a week using a drip system, with the well-watered treatment receiving a total of ∼300 mm over the season (consistent with commercial cotton production) and the water-limited treatment receiving ∼40% to 50% of that quantity (starting later and ending earlier than the well-watered treatment). Other management practices were consistent with commercial cotton production for both irrigation treatments.
Phenotypic Measurements
In year 1, both DM and SC were measured on each F2 plant. At the stage of ∼50% boll opening, dry leaves and branches with open bolls were removed, preventing loss of DM caused by defoliation. At full boll opening, the remaining SC and above-ground plant parts were harvested. In year 2, because within-family replication permitted destructive sampling, one plant per plot was harvested for DM at 50% boll opening, whereas SC was harvested from a second plant at full boll opening. All plant parts were oven dried (except SC, which was air dried) and weighed.
All physiological measurements were conducted during flowering and boll development period using the youngest fully expanded leaf per plant. Although we recognize that measurement of multiple leaves per plant would be ideal, the size of this field experiment and need to sample all plants at similar developmental stages precluded such measurement. Chlorophyll-content analysis was conducted twice during the season: Each time, six leaf disks (with a total area of 1.7 cm2) were sampled from one plant per plot during morning and immersed in 2 mL of N,N-dimethylformamide in the dark for 48 h at 4°C; absorbance of the supernatant at 647 and 664 nm was measured using a spectrophotometer (Uvikon 930, Kontron Instruments), and chlorophyll a and chlorophyll b concentrations were calculated (Moran 1982). Whole-leaf samples were taken at dawn for OP, placed in screw-cap plastic test tubes, frozen in liquid nitrogen, and kept at −18°C until measured. Leaves were defrosted, and OP of the leaf sap was assessed using a vapor-pressure osmometer (5500, Wescor Inc.). Canopy temperature was measured twice during the season at midday with an infrared thermometer (510B, Everest Interscience Inc.). For δ13C analysis, 5-mm (diameter) leaf disks were taken from the youngest fully expanded leaf and each of the two leaves below it at the 30% to 50% boll ripening stage; these were oven-dried, powdered, and combusted in an elemental analyzer (Carlo Erba EA1108, Fison Inc.). The CO2 generated was passed with helium carrier gas directly into the inlet of an isotope ratio mass spectrometer (Opti, Micromass). The 13C/12C ratio was measured and expressed in the δ13C notation relative to the standard Pee Dee Belemnite (δ13C = [Rsample/Rstd − 1]1000, where Rsample and Rstd are the isotope ratios of the sample and standard, respectively).
Cotton fiber was separated from seed using a saw gin. Fiber span length, length uniformity, fineness, strength, elongation, and color were determined with a high-volume instrument (HVI) tester (Zellweger Uster Ag).
Genotyping and Data Analysis
A total of 253 restriction fragment length polymorphism loci spaced at average intervals of 23.1 cM were detected by published procedures using DNA probes sampled from a previously published map (Reinisch et al. 1994), supplemented with new probes (A.H. Paterson, unpubl.) to fill gaps. QTL analyses were performed using Mapmaker-QTL (Lander and Botstein 1989), for a total of 10 data sets, including each of the four individual year × irrigation treatment combinations, two combined across the respective irrigation treatments, two combined across the respective years, one combined across both year and irrigation treatments, and one based on relative values (water limited/well watered) for the replicated year 2 study. A QTL that revealed a LOD difference >2 between environments was considered to show genotype × environment interaction with predominant effect under the treatment with the higher LOD. Most significant interactions showing LOD difference >2 were corroborated by single-point analysis of variance using SAS (Joyner 1985), based on genotype at the nearest single marker(s). As would be expected, single-point analysis of variance missed a few interactions that were detected using interval analysis. Crop performance under stress relative to control is a widely accepted measure of stress adaptation; therefore, QTLs derived from the relative data set were also considered to represent adaptation to water-limited conditions. Based on the length of genetic map and density of markers (above), a LOD = 3 threshold (α = 0.001 on a nominal basis, or 0.05 after accounting for multiple comparisons;Lander and Botstein 1989) was used to declare QTLs. Permutation tests (Churchill and Doerge 1994) were also performed for all traits, generally indicating thresholds between 3.75 and 4.5, and indicating that LOD = 3 corresponded to α ∼ 0.25 to 0.35 (after accounting for multiple comparisons). Higher thresholds were indicated for SC (LOD threshold = 5.03, α = 0.45 for LOD = 3) and for OP (LOD threshold = 6.03, α = 0.64 for LOD = 3); however, for all the QTLs presented in this paper, α < 0.3 based on permutation tests. The hypergeometric probability function (Larsen and Marx 1985) was used to evaluate correspondence between QTLs for different traits, as described (Lin et al. 1995; Paterson et al. 1995). A match was declared when 1-LOD likelihood intervals for two QTLs overlapped.
Acknowledgments
We thank the Paterson, Saranga, and Yakir laboratories for help and encouragement and H. Earl and M. Navarro for valuable comments. We gratefully acknowledge the support of research grant no. US-2506–94R from United States–Israel Binational Agricultural Research and Development (BARD) Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵5 Corresponding author.
-
E-MAIL paterson{at}dogwood.botany.uga.edu; FAX (706) 583-0160.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.157201.
-
- Received July 27, 2000.
- Accepted September 12, 2001.
- Cold Spring Harbor Laboratory Press