A Gene Expression Screen in Zebrafish Embryogenesis
Abstract
A screen for developmentally regulated genes was conducted in the zebrafish, a system offering substantial advantages for the study of the molecular genetics of vertebrate embryogenesis. Clones from a normalized cDNA library from early somitogenesis stages were picked randomly and tested by high-throughput in situ hybridization for restricted expression in at least one of four stages of development. Among 2765 clones that were screened, a total of 347 genes with patterns judged to be restricted were selected. These clones were subjected to partial sequence analysis, allowing recognition of functional motifs in 163 among them. In addition, a portion of the clones were mapped with the aid of the LN54 radiation hybrid panel. The usefulness of the in situ hybridization screening approach is illustrated by describing several new markers for the characteristic structure in the fish embryo named the yolk syncytial layer, and for different regions of the developing brain.
Embryonic development is accompanied by regulated changes in the expression of large sets of genes. Determining how the interplay of these changes influences the progress of development at the cellular and organismic level is a major aim of developmental biology. In the past several decades, it has become clear that the expression and function of a variety of regulatory genes guides developmental processes such as cell differentiation and pattern formation, and it further emerged that a highly effective way of approaching questions of developmental mechanism is to study the properties of differentially regulated gene expression during embryogenesis (Gilbert 2000). This approach has been applied to vertebrate systems in a variety of ways. Earlier studies have emphasized specific aspects of developmental control by selecting genes for study by various criteria, including temporal patterns of expression (Sargent and Dawid 1983), regional restriction (Blumberg et al. 1991), and functional characteristics (Smith and Harland 1991). Such approaches have led to a wealth of information about gene expression patterns providing useful regional markers, and yielding insights into regulatory factors that control differentiation and pattern formation (Cho et al. 1991; Smith and Harland 1992; Sasai et al. 1994; Knecht et al. 1995; Bouwmeester et al. 1996; Richter et al. 1988).
As developmental biology entered the genomic era, the notion has gained currency that it may be not only desirable but also feasible to characterize the regulated expression of the entire population of genes that affect embryogenesis rather than focus on selected subsets of genes. Even in cases where a complete genome sequence is available, this aim is quite large. By placing the focus on those genes whose expression is spatially and temporally regulated during development, however, the total numbers that need to be studied is reduced and the yield of useful information is increased. Screens of this nature have been carried out with Xenopus and mouse embryos, yielding a large selection of genes with highly regulated expression patterns (Gawantka et al. 1998; Neidhardt et al. 2000); a screen in zebrafish carried out by C. Thisse and B. Thisse has been referred to in several publications dealing with individual genes (e.g., Furthauer et al. 2001; Kikuchi et al. 2001). Several types of results can emerge from such a screen. First, interesting expression patterns can lead the investigator to select individual genes for further study (Furthauer et al. 2001; Kudoh and Dawid 2001). Second, genes expressed in similar complex patterns often prove to encode factors that participate in a common signaling or metabolic pathway; such synexpression groups (Niehrs and Pollet 1999) can lead to the discovery of novel components of known pathways (Onichtchouk et al. 1999; Tsang et al. 2000). Third, the availability of numerous novel markers that identify different embryonic domains or cell types facilitates a variety of studies on lineage relationships and developmental functions of these cells (Chin et al. 2000). Fourth, the use of large marker sets contributes to the ambitious goal of a molecular-anatomical atlas of embryogenesis in which domains of gene expression are used to refine and redefine the anatomical descriptions that have classically been used to characterize the embryo.
In this paper, we describe a screen of gene expression patterns in the zebrafish Danio rerio, using high-throughput in situ hybridization with clones derived from a normalized embryonic cDNA library. The zebrafish embryo is an important system for the study of vertebrate embryogenesis, offering advantages that have been expounded in many recent publications. In brief, the high fecundity and small size of this animal, combined with rapid development and the extraordinary optical clarity of the embryo, allow highly effective embryological studies (Kimmel et al. 1990) as well as the execution of large genetic screens (Driever et al. 1996; Haffter et al. 1996;Amsterdam et al. 1999). The benefits of a broad cDNA expression screen are therefore expected to be especially pronounced in the zebrafish. Further, the many mutations available in the zebrafish add a fifth potential use for the products of an expression screen, that is, to provide candidate genes for known mutations. The cloning of genes responsible for a chemically induced mutation is still quite cumbersome, and consequently the candidate gene approach has been very helpful in this context (Schulte-Merker et al. 1994; Talbot et al. 1995; Rebagliati et al. 1998; Sampath et al. 1998; Kikuchi et al. 2001). In addition to an expression pattern that fits the phenotype of a mutation, mapping data are critical in turning a cloned cDNA into a candidate gene. For this reason we have been involved in the establishment of a panel for radiation hybrid (RH) mapping in the zebrafish (Hukriede et al. 1999) and in the extension and refinement of the resulting map (Hukriede et al. 2001). We have used these tools to place a portion of the cDNAs studied on the zebrafish map, enhancing their potential usefulness. In this report, we discuss the methods used in our in situ-based screen and focus on the isolation of marker genes for two important regions of the embryo, the yolk syncytial layer (YSL) and the brain.
RESULTS
Preparation and Evaluation of a Normalized Library
In random screening projects based on cDNA libraries, normalization has proven a useful tool to reduce repetitive analysis of clones representing abundant mRNAs (Soares et al. 1994; Takahashi and Ko 1994). As starting material, a directional cDNA library was prepared from bud to 10-somite-stage embryos in the vector pBluescript KS+, yielding 2 × 106 independent clones with an average insert size of 1.4 kb. This cDNA library was normalized (Bonaldo et al. 1996) with the addition of a sizing step (see Methods), yielding a normalized library containing 2 × 106 clones with an average size of 2.0 kb.
To judge the level of normalization that was achieved, we carried out sequence analysis of groups of randomly selected clones from the original and the normalized library (Table1). Among sequences that allowed interpretation of coding potential, we list four classes of mRNA representing highly abundant protein families. These four classes of abundant products account for 17% of sequenced clones and 36% of identified genes in the original library, and account for 2% of sequenced clones and 6% of identified genes in the normalized library. Therefore, a substantial reduction in the representation of abundant mRNAs was achieved by normalization. This conclusion is supported by the fact that only eight genes were picked more than once during the entire screen.
Representation of Abundant mRNAs in the Parental and Normalized Libraries
Screening for Expression Patterns by In Situ Hybridization
Randomly picked cDNA clones were used to prepare probes for high-throughput in situ hybridization to four stages of zebrafish embryos, as described in the Methods section. Probes that gave staining patterns restricted to some regions of the embryo were analyzed further. Table 2 summarizes the results obtained. The total yield of genes that were judged to have restricted expression patterns was 13%, a proportion that is comparable with the values obtained in the Xenopus screen (15%) and the mouse screen (8%) reported previously (Gawantka et al. 1998; Neidhardt et al. 2000). A broad range of staining patterns was seen, ranging from examples restricted entirely to a single organ, to highly complex patterns in which many but clearly selected tissues express the cognate gene. We subjected each clone to at least a single-pass sequencing run from the 5′ end of the cDNA in the expectation that this would maximize our chances for obtaining coding sequence information and consequently similarity to known protein families or functional motifs. In many, but not all cases; we carried out sequence analysis from the 3′ end as well, and in some cases the entire sequence was obtained. The accession numbers for these sequences are the consecutive numbers from BG985432through BG985856 and are listed explicitly in supplementary Table 3, available at http://www.genome.org.
Summary of Expression Screen
Based on sequence information, close to half of the studied genes either represent previously characterized zebrafish genes or contain known structural motifs that allowed their assignment to protein families (Table 2). The distribution of the 163 genes classified in this way into major functional groups shows that they encode a preponderance of transcription factors or extracellular, transmembrane, and intracellular components of signal transduction cascades (Table 2).
Genomic Mapping of cDNA Clones
The cDNAs characterized in the screen were subjected to mapping by the RH technique, using the LN54 panel (Hukriede et al. 1999, 2001). In this manner, map positions could be assigned to 123 clones whose positions on the LN54 RH map are shown in Figure1, relative to a selected set of previously mapped genes and microsatellite markers.
Map positions of differentially expressed cDNA clones, based on the LN54 mapping panel (Hukriede et al. 1999, 2001). On each linkage group (LG) a selected set of previously mapped genes (in blue) and microsattelite markers (in black) are shown for orientation; clones from the present screen are shown in red.
Spatial and Temporal Complexity of Gene Expression Patterns in the YSL
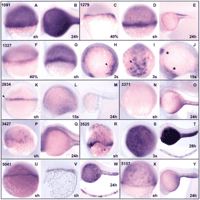
We illustrate the use of cDNA clones from the screen by presenting two examples of embryonic regions whose characterization may be facilitated by the sets of specific markers that have been identified. The YSL is a structure unique to fish that is not found in other vertebrate embryos. Nevertheless, possibly equivalent tissues are found in other animals, such as the yolky endoderm in Xenopus and the visceral endoderm in the mouse. Recently, the dorsal or anterior domains of these three tissues have been shown to act as signaling centers for early dorso-anterior axis formation (Bouwmeester and Leyns 1997; Beddington and Robertson 1998; Solnica-Krezel 1999). Further, its location implies a role in the transfer of nutrients from the yolk to the blastoderm, but this role of the YSL is not fully characterized. In the screen described here, several genes expressed in the YSL were found, substantially increasing the range of markers available for this embryonic domain. Most of these genes were expressed only in the YSL and continued their expression throughout all developmental stages tested. The products of these genes may be structural components of the YSL or have a role in the transfer of materials from the yolk to the blastoderm. The set of genes expressed in the YSL is illustrated in Figure 2. The range of expression patterns includes genes that are expressed strongly throughout the period tested (Fig. 2A,B) and others that are down-regulated after gastrulation (Fig.2K–M). A few of these genes are inactive early, but become expressed by 24 h (Fig. 2N,O). We found only one example of regionally restricted expression within the YSL (Fig. 2F–J).
YSL-specific genes show a variety of expression patterns in embryos from gastrula through the 24-h stage. Each group of panels for one particular clone is enclosed by a black line, with the clone name attop left; stages are shown at bottom right. Clone 1091 has similarity to fructose-1,6-bisphosphatase and is expressed in the YSL at all stages tested (A,B). Clone 1279 is highly homologous to the HNF4 transcription factor. It is expressed in the YSL during early stages (C,D) but not by 24 h (E); expression seen at 24 h is not in the YSL. Clone 1327 has similarity to 4f2/CD98. It is expressed in the entire YSL at early stages (F,G), becomes restricted to posterior YSL at the 3-somite stage (H,I, arrowhead), and expression in the YSL is lost by the 15-somite stage (J); anterior non-YSL expression is indicated by an asterisk. Clone 2834 has similarity to amino acid transporter, solute carrier family 7. It is uniquely expressed in the outer YSL at the shield stage (K, arrowhead). Expression gradually decreases (L) and is lost by 24 h (M). Clone 3371, which is similar to transketolase, is not expressed at gastrula (N) but later becomes expressed in the posterior YSL (O). Clone 3427 is an unidentified sequence that is expressed in the YSL at the shield stage (P) and disappears from the YSL at 24 h but gains a new expression domain in the nervous system (Q). Clone 3525 is similar to, and probably represents, transferrin. It is expressed in the YSL in all stages (R–T); inset in T shows a section, confirming YSL expression. Clone 5041 is an unidentified sequence showing a punctate expression pattern at shield stage (U) and especially at 24 h (W). YSL expression was confirmed by sectioning a shield stage (V) and 24-h embryo (W, inset); the section inV is parasagital and slightly oblique, reducing the thickness of the blastoderm relative to the yolk. Clone 5157 is similar to phosphoenolpyruvate carboxykinase 1 and is expressed in the YSL at all stages tested (X, Y). (40%) 40% epiboly stage; (sh) shield stage; (3s) 3-somite stage; (15s) 15-somite stage; (24h) 24-h stage; (28h) 28-h stage.
The nature of some of the YSL genes could be inferred from sequence comparison. Several of the genes appear to encode metabolic enzymes, such as fructose-1,6-bisphosphatase (clone ID number 1091; Fig. 2A,B), transketolase (3371; Fig. 2N,O), and phosphoenolpyruvate carboxykinase 1 (5157; Fig. 2X,Y). The expression of the iron-transport protein transferrin (3525; Fig. 2R–T) in the YSL is in accord with the apparent transport function of this tissue. Some of the genes that were expressed in the YSL at early stages but disappeared later might have a role in YSL differentiation. Among the genes in this category, clone 1279 (Fig. 2C–E) has high similarity to HNF4. In the mouse, HNF4 as well as transferrin are specifically expressed in visceral endoderm (Meehan et al. 1984; Chen et al. 1994), supporting a view of functional similarity between fish YSL and mouse visceral endoderm not only in the dorsal region that acts as an early organizer, but throughout these tissues. A role for HNF4 in differentiation of this region is supported by the observed defect in visceral endoderm differentiation in mice mutant for the gene (Chen et al. 1994). Clone 1327 likewise is expressed early in the YSL and is subsequently down-regulated in an anterior-to-posterior progression (Fig. 2F–J). This clone is highly similar to human 4f2/CD98, which has been characterized as a cell fusion regulatory protein (Ohgimoto et al. 1995). Because the YSL is formed by cell fusion (Kimmel and Law 1985), it is tempting to speculate that clone 1327 has a role in YSL formation. Many of the genes showed a punctate expression pattern (Fig.2R), which may be apparent only at some stages of development (Fig.2F–I). Yet other genes are expressed continuously throughout the YSL, such as clone 1091/fructose-1,6-bisphosphatase (Fig. 2A,B). These differences appear to depend on the localization of mRNA, in that restriction to a perinuclear domain leads to a punctate pattern, whereas free diffusion of the RNA results in homogeneous staining of the entire YSL domain. The complexity of gene expression patterns within the YSL indicates that this tissue is not homogeneous spatially and that it undergoes considerable differentiation during embryogenesis.
The Complexity of Gene Expression Patterns in the Head
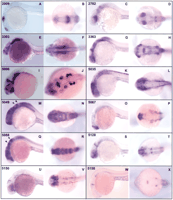
It is well known that the cellular complexity of the brain is associated with a particularly high complexity of gene expression patterns. The characterization of genes that are restricted to certain regions or cell types within the brain has been most helpful in pointing to factors with various functions in this tissue and has also led to a more detailed description of brain structure than otherwise possible. In our screen, we have isolated a number of genes that mark different regions of the brain and more generally, the head. A selection of these genes and the patterns in which they are expressed is presented in Figure 3. The patterns range from highly restricted to more broadly, but still differentially expressed. The former is represented by clone 2009 of unknown nature, which marks the telencephalon and a region within the tectum; and by clone 5158, encoding a KFGF-like molecule, that is limited to a small region in the MHB and a cranial ganglion (Fig. 3, A,B and W,X, respectively). In contrast, clones 5049 (ephrin B3-like) and 5088 (encoding a POU domain protein) are expressed widely in the brain but with low (5049) or high (5088) expression in rhombomeres 3 and 5 (Fig.3, M,N and Q,R, respectively); in addition, clone 5088 is excluded from the telencephalon. Various kinds of regional specialization can be seen, for example the expression of clone 2782 (unknown) in the posterior retina (Fig. 3C,D). These examples illustrate a subset of the wide range of patterns that were observed and point to the potential of using clones from this set in more detailed characterization of the regionalization of the brain.
Head-specific genes at 24- to 28-h stage. For each clone we list the name, sequence similarity, and major expression sites; the left panel gives a lateral view, the right panel an anterior/dorsal view. (A, B) 2009/no similariy/telencephalon, tectum. (C, D) 2782/no similarity/ventral retina, hindbrain. (E, F) 3303/pig10-like/forebrain, lateral hindbrain, otic vesicle. (G, H) 3363/fibronectin-like/forebrain, MH boundary, hindbrain. (I, J) 5006/fgf3/MH boundary, cranial ganglia. (K, L) 5035/no similarity/dorsal diencephalon, MH boundary, row of dorsal neurons. (M,N) 5049/ephrin B3-like/widely expressed except rhombomere 3 and 5 (arrowheads). (O, P) 5067/no similarity/telencephalon. (Q, R) 5088/POU domain/widely expressed except telencephalon, especially strong in rhombomere 3 and 5 (arrowheads). (S, T) 5128/no similarity/restricted regions in fore-, mid-, and hindbrain. (U, V) 5150/IGFBP-like/midbrain. (W,X) 5158/KFGF-like/MH boundary, cranial ganglion.
A Web-Based Database for the cDNA Expression Screen
The data collected from the expression screen have been organized in a database that is accessible on the Web athttp://zf.nichd.nih.gov/pubzf. The site can be searched in several ways. Searching by “clone=” allows a text search using specific gene names or broader terms such as “>homeobox=.” All clones mapped to a particular linkage group can also be displayed, and one can find an entry by its four-digit identification number. A Browse function allows viewing all clones in the database. Searching by “>sequence=” allows the user to paste in a sequence of interest and find database entries by similarity. Perhaps the most useful function in the present context is a search by “>expression pattern=” that can be carried out with the use of a menu of anatomical terms.
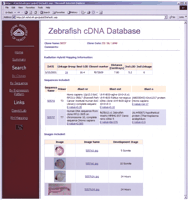
A typical Web page representing one particular clone is shown in Figure4. The mapping information is linked to the LN54 radiation hybrid map (Hukriede et al. 1999, 2001). The results ofBLAST searches with sequence from the clone are listed, providing links to the NCBI Web sites. Finally, in situ hybridization images are shown; clicking on the image name leads to an enlarged and annotated version of the image.
Example of Web page for clone 5057. The page and its links provide the following information: RH-based mapping data; results ofBLAST searches (which in this example reveal close similarity to expressed sequence tags and predicted proteins but not to any functionally characterized protein) and images of expression patterns obtained by in situ hybridization, where clicking on the image name leads to a larger annotated image.
DISCUSSION
General Properties of the Selected Set of Differentially Expressed Genes
The concept that differential gene expression characterizes and, at least to some degree, regulates embryonic development has a long history (for review, see Davidson 1986). Pursuing the characterization of differentially expressed genes has been a highly fruitful approach to the study of developmental mechanisms, complementing the forward genetics approach in an effective way. The set of differentially expressed genes includes markers of terminal cell and tissue differentiation such as globin, troponin, or serum albumin, as well as factors that regulate embryonic patterning and differentiation such as transcription and signaling factors. Although all differentially regulated genes are of use in studying development, it is the regulatory class of genes whose characterization is most instructive. It is therefore encouraging that ∼75% of the genes in our collection encode putative transcription factors or components of signal transduction cascades (Table 2). The high proportion of this class of genes in the set may relate to several factors. One is the selection of genes by restricted expression pattern, which should discriminate against housekeeping genes required in most or all cells. A second point may be the early developmental stages that were used for the generation of the cDNA library, tailbud to 10-somite stage. Genes that contribute to pattern formation, initial specification, and differentiation of tissues are likely to predominate at earlier stages, whereas products of differentiation are more likely to be expressed later. Further, the normalization procedure that was applied is expected to increase the representation of low abundance genes in the library, which would have enriched for tissue specific transcription and signaling factors.
Although the average insert size of 2 kb in our population is satisfactory for a normalized cDNA library (Bonaldo et al. 1996), many of the clones appear to be incomplete. This estimate is based on the experience with a small number of clones that have been selected for more detailed study and the fact that a substantial fraction of the clones do not display a long and convincing open reading frame (ORF). The latter fact clearly limits our ability to recognize structural motifs in the proteins encoded by many of the clones, making the number of recognizable products a definite underestimate.
Determination of the mapping location of a portion of the cDNAs with restricted expression patterns with the aid of the LN54 RH panel in its expanded form (Hukriede et al. 1999, 2001) provided map positions for 123 clones (Fig. 1). This information increases the potential value of the cDNAs as candidate genes for the cloning of zebrafish mutations.
Synexpression Groups
The term “synexpression group” has been used to signify sets of genes that share a similar complex expression pattern (Gawantka et al. 1998). In several cases, such commonality of pattern has indicated function in a common regulatory or metabolic pathway, and can lead to the discovery of novel components of established pathways (Niehrs and Pollet 1999). In our screen, we noticed two clear examples of similar complex patterns that proved to be informative. A clone, eventually identified as nma, is expressed in a pattern essentially identical to that of bmp4 throughout early development. Thenma gene was shown to encode an inhibitory modulator of BMP activity (Tsang et al. 2000), as also shown for the closely relatedXenopus factor BAMBI (Onichtchouk et al. 1999). A second gene that forms a synexpression group with fgf8 and fgf3has been identified and proved to be a modulator of FGF activity (M. Tsang, T. Kudoh, R. Friesel, and I.B. Dawid, unpubl.). It is notable that both examples involve genes that are regulated by a signal transduction cascade and that each gene encodes a product that is able to modulate the activity of the respective signaling pathway. Molecules that can modulate the activity of signal transduction pathways appear to be quite common, although their role in development is only partially understood (Smith and Harland 1992; Sasai et al. 1994;Bouwmeester et al. 1996; Rattner et al. 1997; Massague 1998; Hsieh et al. 1999; Wotton et al. 1999; Kim et al. 2000). These observations support the view that application of the synexpression concept can lead to the discovery of novel components of even well-studied regulatory pathways.
METHODS
Preparation of a Normalized cDNA Library
Total RNA was prepared from bud to 10-somite stage zebrafish embryos using TRIZOL (GIBCO-BRL), and poly-A RNA was isolated by Oligotex (QIAGEN) according to the manufacturer's instructions. Poly-A RNA was converted to cDNA using the SuperScript Plasmid System for cDNA Synthesis and Plasmid Cloning (GIBCO-BRL). Briefly, RNA was reverse-transcribed to first-strand cDNA using SuperScriptII reverse-transcriptase and a tagged oligo-dT primer, which contains several restriction sites including a NotI site (in capital letters): gactagttctagatcgcgatcgcgaGCGGCCGCccttttttttttttttt. Second-strand DNA was synthesized by Escherichia coli DNA polymerase I in combination with E. coli RNAse H and E. coli DNA ligase. Double-stranded cDNA was ligated withSalI adapter and digested with NotI. This procedure produces directional cDNAs containing a SalI site at the 5′ end and a NotI site at the polyA end. These cDNAs were cloned into the SalI/NotI site of pBluescript KS+ and transformed into E. coli Electromax DH10B (GIBCO-BRL) by electroporation. The average size of the inserts in the original cDNA library was 1.4 kb.
A normalized library was prepared from the original library according to Method 4 of Bonaldo et al. (1996) with some modifications. Independent colonies from the original library (107) were grown in semi-solid agar, and amplified plasmid DNA was purified. The plasmid DNA representing the original library was electrophoresed in 0.8% GTG agarose (SeaKem) and the upper half of the DNA distribution was recovered to reduce the proportion of short inserts in the population. The clones were transformed to DH10B, reamplified in semi-solid agar, and DNA was isolated. The plasmid DNA was enzymatically converted to single strands (ss) by ExoIII/GeneII (GIBCO-BRL). An aliquot of the ss-plasmid preparation was used to amplify inserts by PCR using T3/T7 primers to produce “driver” DNA. Another aliquot of the ss-DNA library was hybridized to this driver DNA together with blocking oligonucleotides that prevent interference by vector sequences. Abundant cDNAs hybridize more rapidly and were subsequently removed by hydroxyapatite (HAP) chromatography. The recovered ss-DNA was made partially double-stranded by Sequenase version 2.0 (USB) using ampicillin primer, and transformed into Electromax DH10B cells.
As a preliminary test of normalization efficiency, we amplified one particular cDNA (fgf3) in the PCR step and used it as a driver in place of the PCR product derived from the total cDNA population. The number of colonies obtained from the mock-normalized library was sixfold higher than from the normalized library, showing that a considerable enrichment had been achieved.
In Situ Hybridization Screening
Independent colonies from the libraries mentioned above were cultured and plasmid DNA was purified by the QIAprep Spin Miniprep Kit (QIAGEN). The plasmids were digested with SalI, and Digoxigenin-labeled RNA probe was synthesized by T7 RNA polymerase (Roche). In situ hybridization was performed essentially as described before (Toyama et al. 1995) with the modification that a 24- or 96-well basket device was used together with appropriate plates for incubation and washing steps. Zebrafish embryos at four stages (shield, 3-somite, 15-somite, and 24-h) were analyzed together for each probe.
A Web-Based Laboratory Management Database
Clone information, such as clone name and linkage group, are stored in the database, which also includes the sequence data for each clone and sequencing primers, as well as BLASTN (Altschul et al. 1997), BLAST-EST, and BLASTX reports associated with the sequences. In addition to the BLASTresults from searching GenBank, self-BLASTN results are compiled for detection of duplicate clones within the database. The database also holds data regarding mapping information, images, and descriptions of expression patterns at different embryonic stages. The Web interface allows searching for clones with a given expression pattern and/or sequence. The system provides for on-line uploading of expression images, and permits classification of each image according to expression pattern, using a controlled anatomical vocabulary. Text annotation of images may be entered to supplement the controlled vocabularies. The database also provides full text search capabilities to the BLAST results. The laboratory management database includes reciprocal links to the LN54 RH mapping site (Hukriede et al. 1999, 2001), and includes the scoring vectors and primers associated with each clone.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Acknowledgments
We thank Michael Rebagliati for suggestions on library construction and Elizabeth Laver for help with zebrafish husbandry.
Footnotes
-
↵1 These authors contributed equally to this work.
-
↵3 Corresponding author.
-
E-MAIL idawid{at}nih.gov; FAX (301) 496-0243.
-
Article published on-line before print: Genome Res.,10.1101/gr.209601.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.209601.
-
- Received August 10, 2001.
- Accepted September 12, 2001.
- Cold Spring Harbor Laboratory Press