Acquisition and Metastability of Centromere Identity and Function: Sequence Analysis of a Human Neocentromere
In this issue, Barry and colleagues (Barry et al. 2000) report the sequence of an 80 kb region of euchromatin from human chromosome 10 that can acquire centromeric activity. This new centromere, or neocentromere, drives stable mitotic inheritance once established. Approximately 40 neocentromeres have so far been identified in humans (Warburton et al. 2000). Patients with such neocentromere-containing rearranged chromosomes are heterozygous for the chromosome aberration [marker deletion, or mardel(10)], and so contain homologous loci that are independently inert or fully functional for centromere activity (Voullaire et al. 1993). This study completes a sequence analysis of the neocentromere region (Barry et al. 1999) and investigates what sequence polymorphisms, if any, occur when acquiring neocentromeric activity (Barry and colleagues 2000). We find no evidence for any sequence change, data that strongly support an epigenetic mechanism for neocentromere identity and regulation.
DNA associated with neocentromere activity in mardel(10) (NC DNA) was previously identified by examining the distribution of centromere proteins (primarily centromere proteins CENPs A and C) on stretched chromosomes, relative to the location of regions identified by fluorescence in situ hybridization (du Sart et al. 1997). The restriction map of this 80 kb region of NC DNA was compared to that of homologous non-neocentromeric (HC) DNA from a non-parental source, which demonstrated that no substantial polymorphisms exist between the neocentromere and wild-type genomic library clones (Barry et al. 1999). However, these results are open to the caveat that small changes in primary DNA structure can be causative in centromeric activity, and that these changes are below the resolution of restriction mapping.
Additionally, the entire NC sequence had been determined and analyzed for motifs or presence of repeat DNAs, some of which have been weakly correlated with centromeric activity. The NC sequence was not significantly different from random sequence in regard to satellite DNAs; however, a notable motif of unknown function, AT28, was discovered, and Koch (2000) has discussed its potential contribution to centromeric activity. A superficial structural similarity to alphoid DNA and the centromere of Saccharomyces cerevisiae were enough to implicate AT28 as a potential centromere seed; however, it was not known whether AT28 was unique to the NC DNA, or also present when centromere activity was absent. Given the small size of theSaccharomyces centromere and the ability of single nucleotide mutations to completely disrupt centromere function (Hyman and Sorger 1995), it is not unreasonable to argue that a small region (∼600 bp) can account for the centromere activity in NC. Barry and colleagues (2000) have put this issue to rest by sequencing two additional sources of this same DNA: loci from an unrelated subject (HC DNA) and from the paternal progenitor chromosome (PnC DNA), both of which are inert with respect to centromeric activity. Sequence comparison between the NC DNA and HC DNA showed 370 single nucleotide polymorphisms (SNPs), leading to the possibility that any subset of these SNPs could be correlated with neocentromere activity. However, the sequence of the centromere-inactive PnC progenitor was identical to the NC DNA, including the AT28 region. This clearly and simply rules out any notion that neocentromeric activity relies on these polymorphisms.
This study presents unequivocal evidence for epigenetic regulation of neocentromere activity on mardel(10). Centromere activity clearly maps to this 80 kb region; yet not a single nucleotide differs between it and parental sequence, which shows no neocentromere activity (as assayed by chromosome segregation and localization of twenty centromere-specific factors (Depinet et al. 1997, Saffery et al. 2000). Something other than DNA sequence, such as chromatin structure, must differ between chromosomes of father (PnC) and son (NC), and must be responsible for distinguishing between centromere-on and centromere-off states (Karpen and Allshire 1997; Murphy and Karpen 1998). The persistence of mardel(10)'s centromere, and the absence of centromere activity on normal 10q, shows that the state of centromere activity is stably propagated through the entire cell cycle once it is established. The neocentromere, then, must remain marked throughout the cell cycle, and the mark must be accurately templated to newly synthesized DNA prior to the next S-phase. If we liken the centromere to any other example of epigenetic inheritance, we are left with a wealth of speculative models underlying a potential mark. At one time or another, structural RNA (Clemson et al. 1996), protein localization (Cavalli and Paro 1998), localized protein modification (Ekwall et al. 1997), and covalent DNA modification (Driscoll et al. 1992) have all been suggested as responsible for epigenetic inheritance. Evidence is mounting for the role of proteins such as CENP-A (Vafa et al. 1999), structural RNA such as Xist (Clemson et al. 1996; Willard 1996), or methylation (Ng and Bird 1999) (in non-ecdysozoa) in maintaining stable epigenetic states. Any of these mechanisms could be responsible for the identity of centromeric chromatin. Data fromHomo sapiens (Choo 1997), Drosophila melanogaster(Williams et al. 1998), and Schizosaccharomyces pombe (Steiner and Clarke 1994; Ekwall et al. 1997; Karpen and Allshire 1997) show that in many organisms the centromeres are epigenetically regulated, suggesting a potential universality of mechanism.
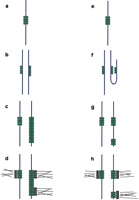
Work on Drosophila has demonstrated that centromere function can spread in cis to juxtaposed DNA (Figure 1a–d) (Williams et al. 1998; Maggert and Karpen,submitted). It is clear that proximity to an active centromere greatly increases the frequency of neocentromere formation on substrate euchromatin, although the mechanism for this is currently unknown. It is unlikely that mardel(10) acquired centromeric activity through this type of spreading. The mardel(10) neocentromere is megabases from the chromosome 10 centromere (Voullaire et al. 1993), and inDrosophila spreading through centric heterochromatin is suppressed (Maggert and Karpen, submitted). However, some chromosome 13 neocentromeres have recently been demonstrated to contain breakpoints near the site of neocentromere formation, suggesting that cis-spreading may be responsible for activation in these examples (Warburton et al. 2000). Spreading of an epigenetic state may also occur in trans, as established for a handful of loci in Zea mays. In one example, the purple plant locus, the frequency of paramutation (or epigenetic mutation) rises in the presence of heterozygous epialleles (Pl-Rh) at the same locus (Martienssen 1996; Hollick 1997), suggesting that epigenetic information can be transferred in trans to homologous loci. Recently, spreading has also been demonstrated forDrosophila dosage compensation (Kelley 1999).
Neocentromeres can arise processively. In (a), a centromere is denoted by association of an epigenetic mark (green circles) with the chromosome (blue lines). In (b) the DNA is replicated, and the mark associates with both chromatids. As the mark is templated to the newly synthesized strand (c), there is some incorporation onto non-centromeric DNA that juxtaposes the centromere. This allows the centromere to increase in size, or to move along the chromosome. Finally, in metaphase (d), kinetochores (blocks and thin lines) are nucleated onto marked DNA, causing dicentric formation on the progenitor to mardel(10). Alternatively, spreading may occur in trans. In (e), the centromere is marked and has sharp boundaries. During or after replication (f), there is a transient interaction between the centromere and an unrelated locus on the same or a different chromosome. (g) Templating of the centromere identity mark assures that this ectopic centromere matures, just as the endogenous centromeres do. Both centromere and neocentromere are capable of nucleating a kinetochore (h), resulting in a dicentric chromosome which undergoes breakage-fusion-bridge during cell division (McClintock 1938).
Although interactions between chromosome regions and spreading in trans are possible (Fig. 1e–h), it is also possible that mardel(10) acquired activity spontaneously. While epigenetic phenomena are generally stable, they also show spontaneous paramutation, as well as reversion, at a rate much higher than that of genetic mutation (Russo et al. 1996). This indicates that although epigenetic states are generally conservative, they can be set or cleared stochastically. If the probability of maintenance of an epigenetic state were orders of magnitude higher than change, one would expect to see long-term conservation of an epigenetic state as well as a low-frequency alteration. This alteration of state, from centromere-off to centromere-on, may explain mardel(10)'s genesis. The rare appearance of neocentromeres in human populations precludes determination of the probability of centromere on-to-off and off-to-on rates. Such experiments have been done in Schizosaccharomyces and show that the frequency of stabilization or deactivation of centromeres are orders of magnitude higher than sequence polymorphisms that underlie more conventional genetic changes (Steiner and Clarke 1994).
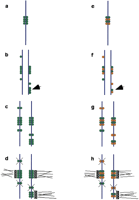
The paradigm of conservative change (metastability) has a model in the bacterial methylase, which is thought to recognize hemi-methylated DNA with a higher affinity than it does unmethyled or fully-methylated DNA (Lewin 1990). Methylated DNA would thus tend to beget methylated DNA after replication, while unmethylated DNA would also tend to remain in that state. With each round of replication, however, some methylated sites could lose covalent modification, and de novo methylation may occur at yet other sites. Figure 2 demonstrates how this metastability can explain the appearance of a neocentromere on mardel(10). Figure 2a shows how a centromere identity factor can interact with any DNA, independent of sequence, defining the location of the centromere. At or shortly after S-phase, identity factor(s) preferentially incorporate like factors to newly replicated DNA at the same locus (Figs. 2b,c), assuring the conserved location of the centromere. But factor-DNA interactions must be stable, and will occur spontaneously, albeit at a much lower frequency, at sites other than the centromere (Fig. 2b). The frequency of these illegitimate events is dictated by the binding affinity of one or more factors for naïve DNA relative to factor-associated DNA. In general, these events will be sporadic, rare, and isolated. Although each interaction has the potential to seed a new centromere, only seeds that fulfil certain criteria mature into centromeres (Fig. 2d). These criteria could be size-dependent (Fig. 2a–d), or sensitive to some other function, perhaps the incorporation of a second self-templating factor (Fig.2e–h). Potential centromeres that do not meet secondary criteria would be wiped clean, perhaps during condensation and kinetochore nucleation or during the next round of replication. This assures a clean slate during subsequent cell divisions. At some very low frequency, one would expect to observe neocentromeres arising in regions where sufficient ectopic incorporation of the identity factor have surpassed the threshold size (Fig. 2a–d) or overlaps with the second factor (Fig. 2e–h). The cell need not monitor the physical size of potential centromeres, but may instead mature the factor-DNA complex at a low frequency, ensuring that longer stretches of identity factor would be more likely to exhibit centromere activity than shorter stretches. Non-centromere DNA is wiped clean, but the activity of the neocentromere preserves itself through cell division. In this model, the dicentric chromosome 10 is subsequently broken and rearranged to generate the mardel(10) and reciprocal rdel(10) chromosomes, though breakage may also have preceded neocentromere formation.
Neocentromeres may arise spontaneously. In (a), a centromere is denoted through interaction of an epigenetic identity mark (green circles) with the chromosome (blue lines). During or after replication (b), the mark is recruited preferentially to DNA that is already associated with the mark, but errors in incorporation yield few sporadic misincorporated marking factors in other parts of the genome. By chance, a local concentration of the centromere mark was incorporated in 10q (arrowhead). These marks are sufficient to recruit more marking factor (c), establishing epigenetically stable potential centromeres. Only potential centromeres of sufficient size (mass of DNA and/or protein) are matured and nucleate kinetochores (d), including the bona fide centromere, as well as the 10q neocentromere. In this case, the active centromeres differ from potential centromeres quantitatively. Alternatively, centromeres may differ qualitatively from potential centromeres. In (e) the centromere is marked by an overlap of more than one centromere identity factor (green and orange circles). Each factor is subject to ectopic incorporation (f). Each factor epigenetically and independently templates the incorporation of like factors (g). In this model, active centromeres are restricted to the subset of potential centromeres where both identity factors are present (arrowhead). The coincidence of centromere identity factors at the bona fide centromere assures that its activity will be stable, and the rare overlap of the two factors defines a neocentromere (h).
The origin of neocentromere activity on mardel(10) is unknown. But the work done by Barry et al. (2000) effectively ends any debate over the epigenetic identity of neocentromeres in human chromosomes. An epigenetic system is one that relies on heritable change without an alteration in DNA sequence, a fact that is now unequivocally established. Mardel(10) is a clear example of this type of epigenetic change, and may serve as a general model for all neocentromere activity. Work from both Dr. Choo's lab and ours suggests that the neocentromere is mechanistically identical to canonical heterochromatic centromeres. As such, mardel(10) represents the first and only metazoan centromere of known and unique sequence. This offers a powerful tool for establishing many aspects of centromere detail. It will be interesting to utilize mardel(10) to understand features of the centromere, such as the extent of the centromere footprint, the identity of the centromere-identity mark, the molecular mechanisms that lead to propagation and spreading, and more.
In this day of whole-genome sequencing, epigenetic phenomena have been underappreciated. Yet, the stable inheritance of states of genome regulation—exemplified recently for the centromere,X-inactivation in Homo sapiens (Clemson et al. 1996), whole-chromosome identification and imprinting inInsecta (Golic 1998; Metz 1938), and mitosis- and meiosis-stable imprinting of gene loci in every kingdom studied (Russo 1996)—may be far from exceptional. If chromosome structure is heavily influenced by epigenetic factors, then it stands to reason that epigenetic alterations would affect chromosome structure. Alterations in gross chromosome structure may be difficult to assay, and may have pleiotropic effects on many aspects of the genome. Such structural requirements may underlie functions that are not easily identifiable by sequence alone. For instance, the difficulty in identifying origins of replication in metazoa, and the identity and function ofDrosophila telomeres (Mason and Biessmann 1995), may be explained by possible epigenetic definition of these structures. Although metazoan origins can be identified in situ, they are typically inactive upon cloning and reintroduction (Françon et al. 1999). Similarly, a broken chromosome end in Drosophila can behave as a double-stranded break in one generation and as a fully-functional telomere in the next, without any alteration in sequence (Mason et al. 1984; Biessmann et al. 1990). These characteristics are reminiscent of epigenetic phenomena; in fact, many chromosomal regulatory features may be epigenetic, including structures necessary for initiation of replication, telomere behavior, gene expression, chromosome identity, chromosome pairing and disjunction, regulation of recombination, and kinetochore nucleation. The lessons that we learn from epigenetic inheritance, and in particular the sequence-independence demonstrated for centromeres, may bear directly on our understanding of many other aspects of chromosome biology.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL karpen{at}salk.edu; FAX 858–622–0417.
- Cold Spring Harbor Laboratory Press