High-Throughput Imaging of Brain Gene Expression
Supplement Figures and Numeric outputs of the algorithms
To plot the figures using the .mat files listed download corresponding figure[1..5].mat files
and run the matlab script plotfigure.m., make sure that figure files are in the same
directory as the script file or that the path variable in Matlab is properly setup.
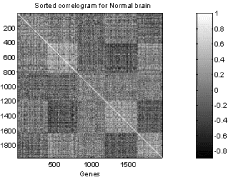

Figure 1 a) : Omnibus correlograms of the two brains
Normal(left) and Alzheimer's(right).
Remark: The correlograms obtained by running the matlab script might have
a different structure which nevertheless should preserve for both images.
Numeric output can be found in the file figure1.mat
|
Variable |
Description |
|
C{1} (matrix, cell array element) |
1977x1977 correlogram of the Normal brain |
|
C{2} (matrix,cell array element) |
1977x1977 correlogram of the Normal brain |
|
reordermin (vector) |
K-means clustering algorithm output (permutation vector) |
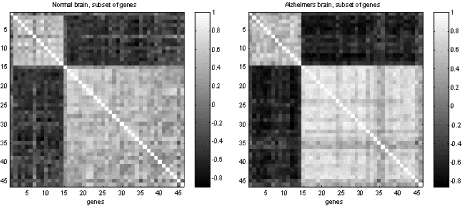
Figure 1 b) : Correlograms of the coregulated genes for the two brains
Numeric output can be found in the file figure1.mat
|
Variable |
Description |
|
C{1} (matrix, cell array element) |
1977x1977 correlogram of the Normal brain |
|
C{2} (matrix, cell array element) |
1977x1977 correlogram of the Normal brain |
|
ips (vector) |
Sorted list of the co-regulated genes, length = 43 elements |
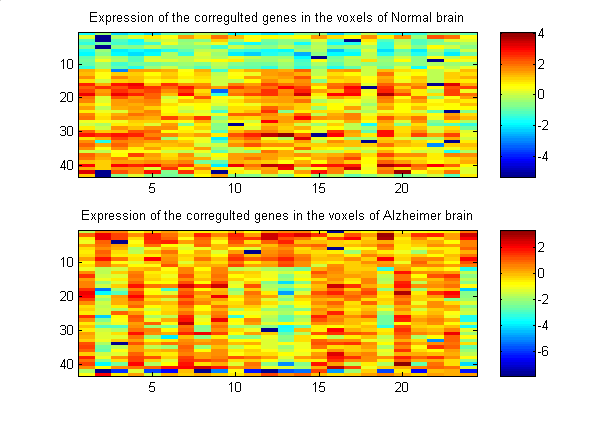
Figure 1 c) : Patterns of expression for the 43 most
differentially expressed genes.
Numeric output can be found in the file figure1.mat
|
Variable |
Description |
|
R{1} (matrix, cell array element) |
1977x24 normalized log-ratio expression levels for 1977 genes in the 24 voxels of the Normal brain |
|
R{2} (matrix, cell array element) |
1977x24 normalized log-ratio expression levels for 1977 genes in the 24 voxels of the AD brain |
|
ips (vector) |
Sorted list of the co-regulated genes, length = 43 elements |
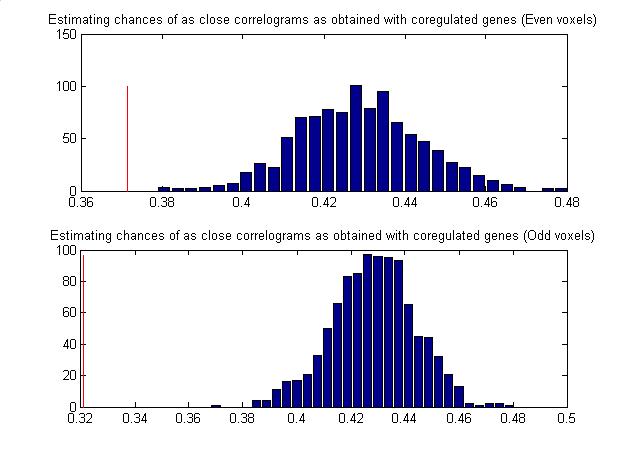
Figure 2) : Estimating chances of the similar correlograms
Numeric output can be found in the file figure2.mat
|
Variable |
Description |
|
evenl (vector) |
Monte-Carlo realizations of the Frobenius norm of the difference between randomly permuted and original correlation matrices of the Normal and AD brains computed with even voxels. |
|
oddl (vector) |
Monte-Carlo realizations of the Frobenius norm of the difference between randomly permuted and original correlation matrices of the Normal and AD brains computed with odd voxels. |
|
Diffipodd (vector) |
Actual value of the Frobenius norm of the difference between correlation matrices of Normal and AD brains computed with even and odd voxels correspondingly. |
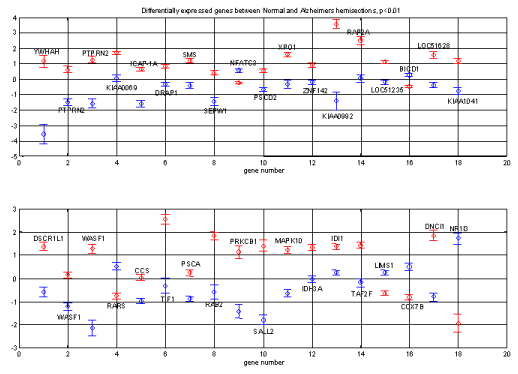
Figure 3 a) : Expression levels for the 40 most differentially expressed
genes between the two brains
Numeric output can be found in the file figure3.mat
|
Variable |
Description |
|
meanR{1} (matrix, cell array element) |
1977x1 normalized log-ratio average expression levels for Normal brain |
|
meanR{2} (matrix, cell array element) |
1977x1 normalized log-ratio average expression levels for AD brain |
|
diffs40 (vector) |
List of the 40 most differentially expressed genes |
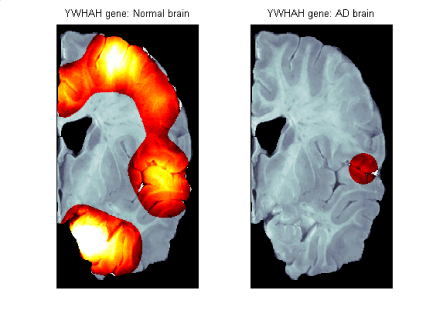
Figure 3 b) : Spatial expression pattern fot YWHAH gene in the
Normal and AD brains
Numeric output can be found in the file figure3.mat
|
Variable |
Description |
|
meanR{1} |
1977x1 normalized log-ratio average expression levels for Normal brain |
|
meanR{2} |
1977x1 normalized log-ratio average expression levels for AD brain |
|
diffs40 (40X1 vector) |
List of the 40 most differentially expressed genes |
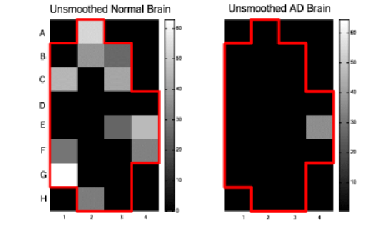
Figure 3 c) : Non smoothed spatial expression patterns for
YWHAH gene in the Normal and AD brains.
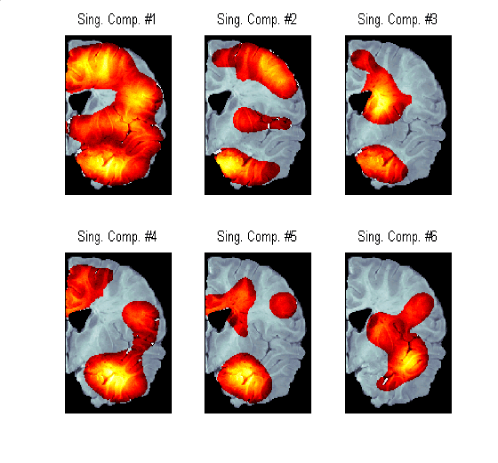
Figure 4) : SVD patterns of the Gene Expression difference between
the Normal and AD brains.
Numeric output can be found in the file figure4.mat
|
Variable |
Description |
|
U (80x24 matrix) |
Matrix of singular vectors of differential expression pattern (gene vectors) |
|
V (24x24 matrix) |
Matrix of singular vectors of differential expression pattern (spatial vector) |
|
S (24x24 diagonal matrix) |
Matrix of singular values |
Follow up experiment. Validation of the presented results
via an independent experiment
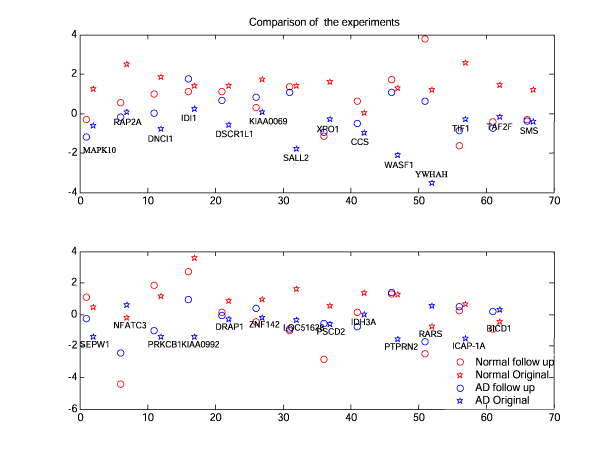
Figure 5 a) : Comparison of the original and follow up experiment data.
Numeric output can be found in the file figure5.mat
|
Variable |
Description |
|
meanR{1} (matrix, cell array element) |
1977x1 normalized log-ratio average expression levels for Normal brain |
|
meanR{2} (matrix, cell array element) |
1977x1 normalized log-ratio average expression levels for AD brain |
|
diffs40 (vector) |
List of the 40 most differentially expressed genes |
|
followupN (31x1 vector) |
Vector of expression levels in the follow-up experiment in the Normal brain |
|
followupA (31x1 vector) |
Vector of expression levels in the follow-up experiment in the AD brain |
|
C (scalar ) |
Correlation coefficient |
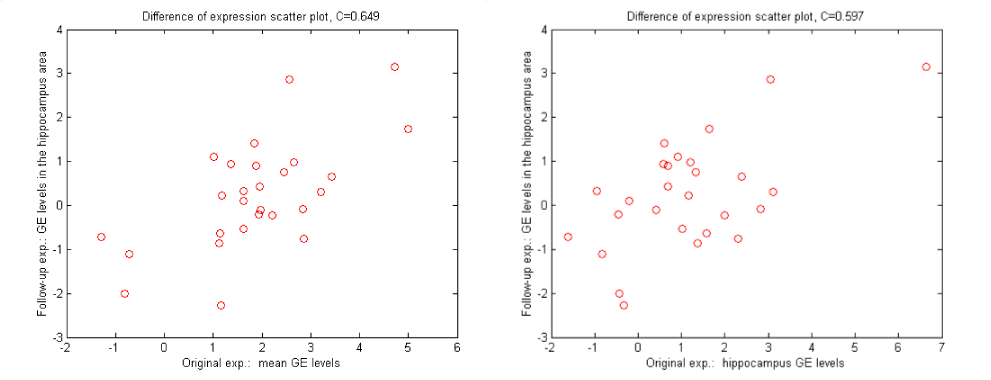
Scatter plots of the gene expression differences.
Left plot is the mean expression in the original experiment vs.
hippocampus in the follow up experiment. C = 0.65
Right plot is the hippocampal expression in the original experiment vs.
hippocampus in the follow up experiment. C = 0.59
This Article
-
doi: 10.1101/gr.204102 Genome Res. February 1, 2002 vol. 12 no. 2 244-254




