Homogeneous Assays for Single-Nucleotide Polymorphism Typing Using AlphaScreen
Beaudet et al.
GR-1725
Supplemental Information
Amplification Controls
To
further improve genotyping accuracy, amplification of an internal control can
be integrated to the AlphaScreen ASA assays to detect PCR failures. An
additional PCR primer pair, which amplifies a 318 bp fragment from the human BGT-1
gene, was included in the PCR/AlphaScreen mix for the WIAF-896 assay. After the
initial reading of the genotypes shown in Figure 1A, specific probes for the BGT-1
PCR product were added to the samples and a second
hybridization step was performed. The results obtained from the reading of a
second 10 ml aliquot are shown in Figure 1B. High
AlphaScreen signal was detected in every well (Fig. 1B), confirming that
PCR/AlphaScreen mix and genomic DNA had been properly dispensed. The addition
of an internal control to the ASA assays will help identify PCR failures, which
were seen in the initial series of ASA, and increase genotyping accuracy.
As with ASA, the most likely
source of errors in genotyping will be from the failure of a sample to amplify.
Thus, a heterozygote could be mistyped as a homozygote. To eliminate this
possible mistyping, two approaches have been tested. First, amplification of an
internal control can be integrated into the assays and detected by the addition
of control probes after the allele-specific detection step, as for the ASA
approach (data not shown). However, a simpler approach has been developed for
ASH, which avoids the extra addition of a second set of probes. Specific probe
hybridization is first performed at the optimized hybridization temperature. An
aliquot of the sample is read and the remaining sample is simply reheated, and
the probes re-annealed at 37°C. At this lower temperature, most probes will
show reduced discrimination for mismatched templates. A significant increase in
signal should be observed for the mismatched allele in homozygous samples if
amplification has occurred. An example of this strategy is shown in Figure 2.
In the first panel, the hybridization step is performed at the optimal
temperature for allele-discrimination (Fig. 2A). Figure 2B shows the results of
the non-specific probe hybridization, made at 37°C. A marked increase in
AlphaScreen signal is observed in the wells of the mismatched alleles for the
homozygous samples, indicating that PCR amplification had indeed occurred in
these wells.
Genotyping from Complex Mixtures
Multiplex
PCR procedures have been developed in several laboratories to save on reagents,
time and operating costs. As a preliminary demonstration of the potential of
AlphaScreen for the detection of targets from multiplexed samples, PCR products
for NCBI SNPs 394, 4568, 4621, 5173 and WIAF-896 were amplified separately from
the same DNA sample and mixed after PCR. The hybridization of the
allele-specific bridging was performed both on the single samples and on an
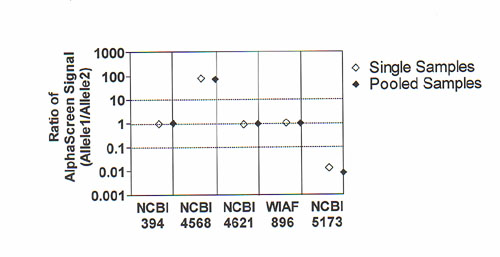
aliquot of the pooled samples. The results are represented graphically in
Figure 3 as a log chart of allelic ratios obtained for each marker. Correct
genotypes were obtained for all the SNP markers and equivalent signal ratios
were observed in both the single and pooled series. These results clearly
indicate that AlphaScreen could be used for the detection of multiplexed PCR
products.
FIGURE
LEGENDS
Figure 1 AlphaScreen ASA genotyping of SNP WIAF-896 in 21 genomic DNA samples. (A) AlphaScreen signal obtained after the detection of the allele-specific amplification product. (B) Detection of the BGT-1 PCR product that was used as a control for PCR amplification. Solid bars, samples amplified with the A-specific PCR primer; white bars, samples amplified with the C-specific primer. Ctrl is a no DNA sample.
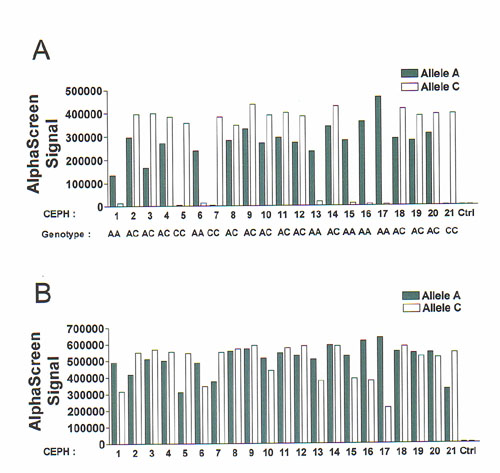
Figure 2 AlphaScreen ASH genotyping assay of SNP WIAF-896 in 21 genomic DNA samples. (A) AlphaScreen signal obtained after allele-specific hybridization of the detection probes at 54.5°C. (B) Control for PCR amplification by the reading of the AlphaScreen signal after probe hybridization at 37°C. Solid bars, samples amplified with the A-specific PCR primer; white bars, samples amplified with the C-specific primer. Ctrl is a no DNA sample.
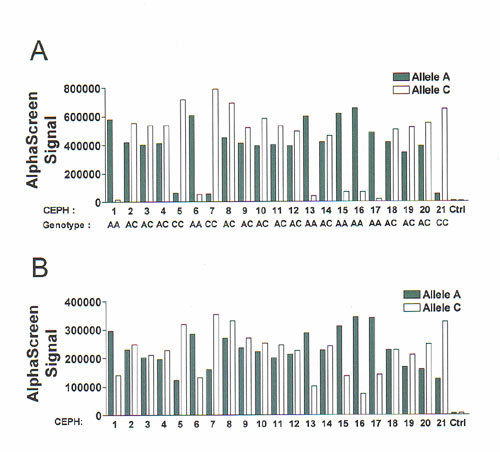
Figure 3 Genotyping of a DNA sample using five different AlphaScreen ASH SNP assays from single or pooled PCR products. PCR was performed for each SNP assay separately. A fraction of the PCR products were pooled together to mimic multiplexed amplification, and split into five for the individual detection of each SNP genotype. The ratio of AlphaScreen signal obtained for the two alleles of each SNP was determined. (¯) AlphaScreen genotyping using the single PCR products; (¿) AlphaScreen genotyping using the pooled PCR products.
This Article
-
doi: 10.1101/gr.1725501 Genome Res. April 1, 2001 vol. 11 no. 4 600-608




