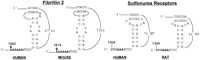Identification of Putative Programmed −1 Ribosomal Frameshift Signals in Large DNA Databases
- 1Department of Molecular Genetics and Microbiology, University of Medicine and Dentistry of New Jersey (UMDNJ), Robert Wood Johnson Medical School, and The Graduate Programs in Molecular Bioscience Rutgers/UMDNJ, Piscataway, New Jersey 08854 USA; 2The Cancer Institute of New Jersey, and 3Molecular Statistics and Bioinformatics Section, Biometric Research Branch (BRB), Cancer Therapy Evaluation Program (CTEP), Division of Cancer Treatment and Diagnosis (DCTD), National Cancer Institute, Rockville, Maryland 20892 USA
Abstract
The cis-acting elements that promote efficient ribosomal frameshifting in the −1 (5′) direction have been well characterized in several viral systems. Results from many studies have convincingly demonstrated that the basic molecular mechanisms governing programmed −1 ribosomal frameshifting are almost identical from yeast to humans. We are interested in testing the hypothesis that programmed −1 ribosomal frameshifting can be used to control cellular gene expression. Toward this end, a computer program was designed to search large DNA databases for consensus −1 ribosomal frameshift signals. The results demonstrated that consensus programmed −1 ribosomal frameshift signals can be identified in a substantial number of chromosomally encoded mRNAs and that they occur with frequencies from two- to sixfold greater than random in all of the databases searched. A preliminary survey of the databases resulting from the computer searches found that consensus frameshift signals are present in at least 21 homologous genes from different species, 2 of which are nearly identical, suggesting evolutionary conservation of function. We show that four previously described missense alleles of genes that are linked to human diseases would disrupt putative programmed −1 ribosomal frameshift signals, suggesting that the frameshift signal may be involved in the normal expression of these genes. We also demonstrate that signals found in the yeastRAS1 and the human CCR5 genes were able to promote significant levels of programmed −1 ribosomal frameshifting. The significance of these frameshifting signals in controlling gene expression is not known, however.
Although maintenance of correct reading frame is fundamental to the integrity of the translation process and, ultimately, to cell growth and viability, an increasing number of cases have been described in which translating ribosomes are intentionally directed to shift reading frame. The great majority of “programmed ribosomal frameshift” events have been observed in RNA viruses (for reviews, see Brierley 1995; Dinman 1995; Gesteland and Atkins 1996; Dinman et al. 1998). In mammals, families of viruses that are known to use programmed ribosomal frameshifting include retroviruses, coronaviruses, toroviruses, arteriviruses, astroviruses, and at least one example in a paramyxovirius. Plant viruses that use this mechanism include tetraviruses and tombusviruses. In fungi, ribosomal frameshifting is used by the totiviruses and many of the retrotransposable elements. Programmed ribosomal frameshifting has been documented in T7 and λ bacteriophages as well (Condron et al. 1991; Levin et al. 1993). Viral frameshifting events typically produce fusion proteins in which the amino- and carboxy-terminal domains are encoded by two distinct, overlapping open reading frames. Ribosomal frameshifting in viruses determines the stoichiometric ratio of structural (Gag) to enzymatic (Gag–Pol) proteins and plays a critical role in viral particle assembly (Felsenstein and Goff 1988; Xu and Boeke 1990; Park and Morrow 1991; Dinman and Wickner 1992, 1994; Karacostas et al. 1993; Kawakami et al. 1993; Cui et al. 1996; Dinman and Kinzy 1997; Dinman et al. 1997; Tumer et al. 1998). The study of these ribosomal frameshifts has been important both because of their critical role in viral particle morphogenesis and because of the information they provide about the mechanisms by which reading frame is normally maintained (for review, see Brierley 1995; Dinman 1995; Farabaugh 1996; Weng et al. 1997;Dinman et al. 1998).
There are a few documented examples in which “translational recoding” (including programmed ribosomal frameshifting and nonsense suppression) is used to control the expression of cellular mRNAs. Translational readthrough of a termination codon has been documented in the kelch (Xue and Cooley 1993), oaf (Bergstrom et al. 1995), and hdc (Stenberg et al. 1998) transcripts ofDrosophila. In Escherichia coli, autoregulation of a programmed +1 ribosomal frameshift in the prfB gene is required for the synthesis of release factor 2 (RF2) (Craigen et al. 1985; Craigen and Caskey 1986; Donly et al. 1990a,b), and a programmed −1 ribosomal frameshift in the dnaX gene generates the DNA polymerase γ-subunit (Blinkowa and Walker 1990; Tsuchihashi and Kornberg 1990; Flower and McHenry 1991). In eukaryotic mRNAs, programmed +1 ribosomal frameshifting has been demonstrated in genes encoding ornithine decarboxylase (ODC) antizyme isolated from human, rat, mouse, Xenopus, and Drosophila (Rom and Kahana 1994; Hayashi and Murakami 1995; Ichiba et al. 1995; Matsufuji et al. 1995; Kankare et al. 1997; Ivanov et al. 1998a,b) and in theEST3 gene of Saccharomyces cerevisiae (Lundblad and Morris 1997). In mammalian cells, the control of ribosomal frameshifting efficiency is autoregulated by ODC antizyme protein levels (Hayashi and Murakami 1995; Matsufuji et al. 1995). Thus, the regulation of polyamine biosynthesis demonstrates how programmed ribosomal frameshifting may be used by eukaryotic cellular genes as a post-transcriptional control mechanism.
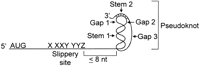
Although there are no known reported examples of eukaryotic cellular mRNAs that use programmed −1 ribosomal frameshifting to control protein expression, the cis-acting sequences that promote efficient programmed −1 ribosomal frameshifting have been well characterized in several eukaryotic viral systems (for reviews, seeBrierley 1995; Dinman 1995; Gesteland and Atkins 1996; Farabaugh 1997;Dinman et al. 1998). In eukaryotic viruses, two basic sequence elements are required to promote efficient levels of programmed −1 ribosomal frameshifting. The first sequence element is called the “slippery site” and consists of a heptamer sequence X XXY YYZ (the incoming 0-frame, e.g., the gag reading frame, is indicated by spaces), in which XXX can be any three identical nucleotides, YYY can be AAA or UUU, and Z is A, U, or C (Fig. 1) (Jacks and Varmus 1985; Dinman et al. 1991; Brierley et al. 1992; Dinman and Wickner 1992). The second promoting element is usually a sequence that forms a defined RNA secondary structure, such as an RNA pseudoknot. This is located within 8 nucleotides 3′ of the slippery site and is thought to increase the probability that the ribosome will slip reading frame in the −1 direction (Fig. 1) (Tu et al. 1992; Somogyi et al. 1993). The simultaneous slippage of both ribosome-bound tRNAs by 1 base in the 5′ direction still leaves their nonwobble bases correctly paired with the mRNA in the new reading frame. It has been convincingly demonstrated that the basic molecular mechanisms governing programmed −1 ribosomal frameshifting are identical from yeast to humans (Wilson et al. 1988; Dinman et al. 1991; Dinman and Wickner 1992; Stahl et al. 1995).
A consensus programmed −1 ribosomal frameshift signal. The incoming reading frame with regard to the translational start site is denoted by spaces. The distances between slippery site and pseudoknot, the two stems, and the three gaps are defined in Methods.
The general sequence and structural requirements are well enough defined to begin to identify putative programmed −1 ribosomal frameshift sites from the sequences in the large DNA databases. We have constructed a computer program designed to search large DNA databases for consensus −1 ribosomal frameshift signals using an algorithm that is both stringent in its description of the slippery site and its requirement for a general pseudoknot structure but that is liberal with regard to the spacing between the slippery site and pseudoknot and with regard to specific G + C content of the first half of stem 1. The specific parameters were chosen to maximize the probability of finding sequence elements that have a reasonable chance of promoting ribosomes to shift reading frame with frequencies that are significantly greater than background. The results of our analyses show (1) that consensus frameshift signals occur at frequencies significantly greater than random, (2) that some frameshift signals appear to be evolutionarily conserved between homologous genes in different species, and (3) that mutations that have been linked to inherited human diseases correlate with those that are predicted to abolish programmed −1 ribosomal frameshifting. Furthermore, we demonstrate that at least two of the signals identified by the program are able to promote significant levels of programmed −1 ribosomal frameshifting in a yeast assay system. At present, the role of programmed frameshifting in controlling expression of these mRNAs is not known. Based on these results, the potential role of programmed −1 ribosomal frameshifting or the role of the frameshift signal in controlling translation efficiencies and mRNA turnover are discussed.
RESULTS
Development of a Computer Program Capable of Finding Known Viral Programmed −1 Ribosomal Frameshift Signals
The primary objective of this study was to use a computer search protocol to identify sequence elements that have a reasonable chance of promoting ribosomes to shift reading frame with frequencies that are significantly greater than background in the large DNA databases (see Methods). To determine whether the computer program was capable of properly identifying known frameshift signals, we conducted a search of all 36,556 loci of the GenBank virus division. The results of this search revealed 1077 motif hits from among the 3.7 × 107base pairs in the database (Table 1). The program identified almost all of the known viral −1 ribosomal frameshift signals including those that have been classically used to study programmed −1 ribosomal frameshifting. These include mouse mammary tumor virus, barley yellow dwarf virus, and infectious bronchitis virus. As expected, the program was not able to identify the motif hit in Rous sarcoma virus (RSV) because gaps 1 and 2 are larger than allowed by the program. Interestingly, many motif hits were identified in families of viruses in which −1 ribosomal frameshifting has not been described (data not shown).
Summary of Search Results
Consensus Motif Hits Occur at Frequencies Significantly Greater Than Random in the Genome Databases
We then addressed the question of whether consensus programmed −1 ribosomal frameshift signals can be found in the large DNA databases at frequencies significantly greater than random. To test this, we first determined the probability of the random occurrence of a motif hit. A zero-order Markov model consisting of two sets of 104randomly generated sequences composed of 103 bases each (50% G + C content) was chosen as the negative control set. This model was chosen based on the reasoning that if programmed −1 ribosomal frameshifting does not have a function in a subset of chromosomally encoded mRNAs, then consensus frameshift signals should be randomly distributed throughout genomes independently of any nearest neighbors. Thus, the collection of negative control sequences that were used represents the true null set, and comparisons that arise from this control should be meaningful. The program found 41 motif hits in the first set of random sequences and 42 in the second set. Thus, the random frequency of motif hits is 83 per 2 × 107 base pairs (Table 1).
Having empirically established the random frequency of motif hits, the computer program was then applied to the large DNA databases. These searches revealed that motif hits occur with frequencies approximately two- to sixfold more frequently than random (Table 1). Analysis of theS. cerevisiae data set revealed 260 motif hits, ∼5.2-fold more frequent than random. BLAST analysis revealed that 150 different recognized genes or CDS were represented in the motif hits. Because the yeast genome is estimated to contain ∼5900 genes (seehttp://genome-www.stanford.edu/Saccharomyces/), these data suggest that at least 2.54% of the genes in the yeast genome contain at least one consensus programmed −1 ribosomal frameshift signal.
Frameshift Signals Appear to be Evolutionarily Conserved Between Homologous Genes in Different Species
If the frameshift signal is used to control the expression of a subset of cellular mRNAs, then we predict that specific frameshift signals should be evolutionarily conserved in homologous genes from different organisms. A preliminary comparison of the locations and structures of motif hits in homologous genes in the different databases revealed at least 21 homologous genes from different species that contained consensus frameshift signals (Table 2). In two such cases, the Fibrillin 2 genes of human and mouse and the Sulfonurea receptor genes of human and rat, the frameshift signals are nearly identical (Fig. 2).
Frameshift Signal Containing Homologous Genes from Different Species
Comparison of the human and mouse Fibrillin 2 and the human and rat Sulfonurea receptor motif hits. Numbers refer to the 5′ and 3′ positions of the depicted nucleotides in the respective ORFs. Slippery sites are italicized in boldface.
Mutations that Have Been Linked to Inherited Human Diseases Correlate with Those That Are Predicted to Abolish −1 Ribosomal Frameshifting
If the frameshifting signal has a biologically relevant function in cellular gene expression, then we should be able to correlate mutations that disrupt the frameshifting with demonstrable phenotypes. One place to look for phenotypes that correlate with mutations that disrupt putative frameshift signals is among certain of the well-characterized genetically inherited diseases of humans. Ideally, such alleles would encode simple silent or missense mutations or would either add or delete entire codons in the putative frameshift signal. Such mutations would minimally impact on the primary informaprimary information encoded by their mRNAs, encoding at most a single amino acid change and leaving reading frames intact. They would, however, disrupt the frameshift signal, ablating its ability to make ribosomes shift reading frame. A preliminary analysis of the human motif hit database identified four alleles of three genes that fit these criteria (Table3). In the human gene encoding triacylglycerol lipase, the .0027 allelic variant of triacylglycerol lipase (linked to lipoprotein lipase deficiency) (Wilson et al. 1993) and the .0021 allelic variant (linked to Familial chylomicronemia syndrome) (Gotoda et al. 1992) are both predicted to disrupt the RNA pseudoknot component of the consensus −1 ribosomal frameshift signal. Similarly, the .0007 allelic variant of the FASL antigen (linked to autoimmune lymphoproliferative syndrome) (Bettinardi et al. 1997) is also predicted to disrupt the RNA pseudoknot. Disruption of the mRNA pseudoknot is predicted to abolish programmed −1 ribosomal frameshifting (for reviews, see TenDam et al. 1990; Brierley 1995;Dinman 1995; Farabaugh 1996; Gesteland and Atkins 1996; Jacks 1996;Dinman et al. 1998). In addition, the .0004 allele of the ETFA-electron transfer flavoprotein α-subunit precursor (linked to type II glutaricaciduria) (Freneaux et al. 1992) disrupts the spacing between the slippery site and the RNA pseudoknot, which is predicted to result in a decrease in programmed −1 ribosomal frameshifting efficiency (Brierley et al. 1991, 1992; Dinman and Wickner 1992; Morikawa and Bishop 1992).
Three Human Genes in Which Specific Mutations in the Consensus−1 Ribosomal Frameshifting Signals Have Been Linked to Disease
Computer Identified Motif Hits Can Promote Efficient Levels of Programmed −1 Ribosomal Frameshifting in S. cerevisiae
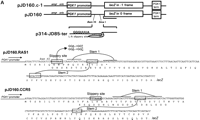
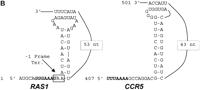
Using a series of frameshift reporter plasmids and yeast strains previously developed in our laboratory (Dinman et al. 1997), we tested whether the yeast RAS1 and human CCR5 motif hits were able to promote efficient levels of programmed −1 ribosomal frameshifting in intact yeast cells. The RAS1 motif hit was amplified by PCR from yeast genomic DNA and was cloned into pJD160. Two additional C residues were added between the slippery site and pseudoknot so that a programmed −1 ribosomal frameshift would be required for translation of the lacZ gene (see Fig.3A). The CCR5 motif hit was similarly amplified from a cDNA clone (see Fig. 3A). This set constitutes the frameshift test plasmids. As a positive control, the efficiency of programmed −1 ribosomal frameshifting as promoted by the L-A virus frameshift signal was determined to compare the frameshift promoting abilities of the motif hits to a known programmed −1 ribosomal frameshift signal. β-Galactosidase activities generated from cells harboring pJD160.c-1 were monitored as a negative control to determine the background levels of nonprogrammed −1 frame-shifting. Ribosomal frameshift efficiencies were calculated by dividing the β-galactosidase activities generated from cells harboring frameshift test plasmids by the β-galactosidase activity generated by the 0-frame control, pJD160. The results of these experiments demonstrate that the RAS1motif hit promoted programmed −1 ribosomal frameshifting with an efficiency of ∼4.4% and that CCR5 promoted a 0.2% efficiency of promoted programmed −1 ribosomal frameshifting (Table4). The L-A signal promoted programmed −1 ribosomal frameshifting with an efficiency of 1.9%, and nonprogrammed frameshifting was <0.01% (Table 4) . These results demonstrate that both the RAS1 and the CCR5 motif hits are capable of promoting programmed −1 ribosomal frameshifting with efficiencies of 4.4% and 0.2%, respectively. These frequencies are >440- and 20-fold greater than background, respectively.
(A) Plasmids used to measure programmed −1 ribosomal frameshifting. pJD160.0 is the 0-frame control plasmid. pJD160.c-1 measures nonprogrammed −1 ribosomal frameshifting. p314-JD85-ter is used to measure L-A virus-promoted programmed −1 ribosomal frameshifting. The frameshift signal from the yeast RAS1 gene was cloned into pJD160 to produce pJD160.RAS1. Because theRAS1 frameshift signal is predicted to direct ribosomes into premature termination signals, two additional nucleotides were added in the spacer regions between the slippery site and pseudoknot of theRAS1 PCR product. The CCR5 frameshift signal was cloned from a CCR5 cDNA template into pJD160.0 to produce pJD160.CCR5. In each of these constructs, a programmed −1 frameshift is required for the lacZ gene to be translated. Ribosomal frameshift efficiencies were calculated by dividing β-galactosidase activities from cells harboring test plasmids by β-galactosidase activities from cells harboring the 0-frame control plasmid and multiplying by 100%. (B) Representations of the RAS1and CCR5 motif hits. Numbers refer to the 5′ and 3′ positions of the depicted nucleotides in the respective ORFs. Slippery sites are italicized in boldface. The RAS1 −1 frame termination codon is noted.
The RAS1 and CCR5 Motif Hits Can Promote Efficient Levels of Programmed −1 Ribosomal Frameshifting in Intact Yeast Cells
Programmed Ribosomal Frameshifting Does not ControlRAS1 Expression
We have shown that the RAS1 motif hit promotes efficient programmed −1 ribosomal frameshifting. We then tested whether ribosomal frameshifting has a role in controlling RAS1 gene expression. We obtained a yeast strain in which both copies ofRAS (RAS1 and RAS2) were deleted. These were then manipulated so that the source of their Ras1 proteins was derived from a set of single copy plasmids harboring either the wild-typeRAS1 gene (pRAS1–TRP1) or RAS1 genes harboring mutations that were silent with respect to their protein coding functions (the pRAS1A → C and pRAS1A → T mutants; see Fig.3A) but that were predicted to be unable to promote efficient programmed −1 ribosomal frameshifting as a consequence of disruption of the slippery site (Jacks et al. 1988; Dinman et al. 1991; Brierley et al. 1992; Dinman and Wickner 1992). If programmed −1 ribosomal frameshifting is the mechanism responsible for the previously observed differences between RAS1 and RAS2 (Breviario et al. 1986), then cells harboring the mutant RAS1 alleles should have been able to use poor carbon sources at 37°C. However, we observed that none of the RAS1 alleles supported cell growth using ethanol, glycerol, or acetate at the nonpermissive temperature (data not shown). Similarly, no differences were observed in the growth rates of cells harboring the wild-type as compared with cells harboring the mutant RAS1 alleles at 30°C, irrespective of carbon source.
DISCUSSION
The goal of this research has been to determine whether programmed −1 ribosomal frameshifting is used by a subset of chromosomally encoded eukaryotic mRNAs. As a first step towards this end, we constructed a computer program based on an algorithm describing a set of consensus programmed −1 ribosomal frameshift signals. The algorithm was structured to allow the identification of sequence elements that have a reasonable chance of promoting ribosomes to shift reading frame with frequencies that are significantly greater than background in the large DNA databases. To this end, the sequence parameters describing the slippery site, the absolute requirement for a pseudoknot structure, and the maximum sizes of loops 1 and 2 were fairly stringent. The limitations placed on other parameters were more liberal, however, to acknowledge (1) previously observed variability between different programmed −1 ribosomal frameshift signals (for review, see Brierley 1995; Farabaugh 1996; Gesteland and Atkins 1996;Jacks 1996) and (2) currently unresolved issues in the field. For example, although the spacing between the slippery site and the pseudoknot ranges from 5 to 8 nucleotides in most viral frameshift signals, known exceptions to this rule [e.g., the spacing in Rous sarcoma virus is only 1 nucleotide (Marczinke et al. 1998)] led us to leave this variable rather broad. Similarly, although many viral frameshift signals appear to require a large number of G + C residues at the start of the 5′ arm of stem 1 (for review, see Farabaugh 1997), this area of stem in some other viruses (e.g., L-A) is not highly G-C rich (Dinman and Wickner 1992). Furthermore, because the precise secondary structural requirements of frameshift-promoting pseudoknots are controversial [e.g., the MMTV pseudoknot appears to require a 112° bend at the interface between stems 1 and 2 (Chen et al. 1995), stems 1 and 2 of the IBV pseudoknot appear to stack coaxially (Brierley et al. 1991), and the secondary structural requirements of the RSV pseudoknot are completely novel (Marczinke et al. 1998)], we chose not to place stringent constraints on parameters that would describe RNA pseudoknot structures. Additionally, there are numerous examples where changes as little as two- to threefold have significant biological impacts. Thus, although viral signals have evolved to promote programmed −1 ribosomal frameshifting with extraordinarily high efficiencies [100-fold or greater than baseline rates of unprogrammed frameshifting (Dinman et al. 1991)], the requirements for the lengths and compositions of stems 1 and 2 and loop 3 were allowed to be less stringent so as to allow the program to identify motif hits that, although less efficient than viral fameshift signals, were potentially capable of promoting programmed −1 ribosomal frameshifting with efficiencies significantly higher than background. In light of the CCR5 results, this approach can be considered successful.
With these aims in mind, we have demonstrated that the program is capable of finding known viral frameshift signals, and we have shown that consensus programmed −1 ribosomal frameshift signals occur with frequencies that are significantly greater than random in the large DNA databases. The results from the S. cerevisiae genome most likely provide the best estimate of the frequency of motif hits, because (1) it is complete, (2) it is on the same order of magnitude as the random control, (3) it contains the least amount of duplications, and (4) it was sequenced without reading-frame bias. In contrast, for example, the large number of sequences derived from expressed sequence tags (ESTs) in the human genome database tend to inflate the total number of nonmotif hit-containing sequences, decreasing the apparent frequency of motif hits in this database. Additionally, because our algorithm limited the size of gaps 1 and 2 and disallowed slippery sites of TTTTTTT and AAAAAAA, our data probably represent an underestimate of the fraction of yeast genes containing consensus programmed −1 ribosomal frameshift signals.
A preliminary comparative analysis of consensus programmed −1 ribosomal frameshift signals from different species’ DNA databases showed that many homologous genes contained motif hits (Table 3) and that almost identical motif hits appear to be evolutionarily conserved in at least two cases (Fig. 2). It is notable that whereas the slippery sites and stems of the frameshift signals in both the Fibrillin 2 and the Sulfonurea receptor mRNAs are highly conserved, the lengths of gap3 which are not expected to play a critical role in frameshifting (Brierley et al. 1989, 1991), are variable in both of these examples. Thus, it appears that the biologically important elements of the frameshift signals have been conserved, whereas the unimportant elements have been allowed to drift.
We have used a yeast-based reporter system to demonstrate that at least two of the motif hits that were identified by the computer program can promote programmed −1 ribosomal frameshifting at levels that are significantly greater than background (Table 4). The 4.4% efficiency of programmed −1 ribosomal frameshifting promoted by theRAS1 motif hit is comparable to frameshift efficiencies promoted by naturally occurring viral frameshift signals (for reviews, see Brierley 1995; Farabaugh 1997). One potential caveat with this measurement, however, is the fact that we had to alter the distance between the slippery site and RNA pseudoknot by adding two C residues to measure β-galactosidase activity and, hence, frameshifting using the enzymatic reporter system. Because changes in this spacing can have dramatic effects on frameshift efficiencies (Dinman and Wickner 1992;Morikawa and Bishop 1992; Kollmus et al. 1994), 4.4% is probably not the actual frameshift efficiency promoted by the native RAS1motif hit.
Although programmed −1 ribosomal frameshifting has heretofore only been observed in some RNA viruses, we hypothesize that this mechanism may also be used to control the expression of a subset of some chromosomally encoded eukaryotic mRNAs. The finding that known missense alleles that are linked to human diseases that are predicted to disrupt the ability of motif hits to promote efficient levels of programmed −1 ribosomal frameshifting and that also minimally impact the primary coding sequence provides circumstantial support for this hypothesis (Table 3). As a first attempt to directly test this premise, we chose to examine the RAS1 motif hit. It has been shown thatras2 mutants are unable to grow on nonfermentable carbon sources at 37°C and that the steady-state level of Ras1 mRNA and the rate of Ras1 protein synthesis are reduced as compared with the Ras2 mRNA and Ras2 protein (Fraenkel 1985; Tatchell et al. 1985; Breviario et al. 1986, 1988). The observed phenotypic differences betweenras1 and ras2 mutants depend on the highly conserved 5′ halves or amino termini of the RAS1 or RAS2genes or their respective proteins rather than on their highly divergent 3′ halves or carboxy-terminal regions (Hurwitz et al. 1995). Because only RAS1 contains the putative −1 ribosomal frameshift signal, we hypothesized that programmed ribosomal frameshifting was responsible for the different phenotypes. Because theRAS1 frameshift signal would cause a shifted ribosome to encounter a premature termination codon, we postulated that frameshift events promoted by this element would activate the nonsense-mediated mRNA decay pathway, resulting in destabilization of the Ras1 mRNA (for reviews, see Maquat 1995; Caponigro and Parker 1996; Ruiz-Echevarria et al. 1996; Weng et al. 1997). By this model, inactivation of the frameshift signal would serve to stabilize the Ras1 mRNA. This would in turn increase Ras1p abundance, allowing the mutants to use nonfermentable carbon sources in the absence of a functionalRAS2 gene. When tested, however, ras2 strains harboring RAS1 alleles with inactivated slippery sites were still unable to efficiently use nonfermentable carbon sources. A potential problem with the RAS1 motif hit may be that its imminence to the translational start site may actually inhibit frameshifting (Belcourt and Farabaugh 1990). The presence of the predicted RNA pseudoknot so close to the translational start site may inhibit translation initiation, which may explain the differences between RAS1 and RAS2.
In sum, we have developed a computer program that is capable of identifying consensus programmed −1 ribosomal frameshift signals in the large DNA databases. We have demonstrated that the program is capable of identifying known viral frameshift signals, we have empirically determined the random frequency of motif hits, and we have demonstrated that these signals occur with frequencies that are significantly greater than random in the large DNA databases. We have also presented indirect evidence that many such signals are evolutionarily conserved and that disruption of putative frameshift signals may be linked with some genetically inherited human diseases. At least two of the motif hits that were identified by the computer program can promote efficient levels of programmed −1 ribosomal frameshifting. Although we were not able to provide direct proof that programmed −1 ribosomal frameshifting controls RAS1expression, the data presented here represent an important first step toward identifying the role that programmed −1 ribosomal frameshifting or the frameshift signal may play as a post-transcriptional control mechanism in the expression of certain cellular mRNAs. Future experiments will focus on identifying transcripts in which programmed −1 ribosomal frameshifting plays such a role.
METHODS
Computer Search Protocols
The GenBank Saccharomyces cerevisiae, Homo sapiens, Mus musculus, Rattus norvegicus, Gallus gallus, Sus scrofa, Drosophila melanogaster, and virus divisions, and 2 × 104random sequences of 103 bases (GC content = 50%) were searched using the following algorithmic structure:
Step 1: Search for XXXYYYZ (slippery site) in which
XXX = GGG, AAA, TTT, or CCC;
YYY = AAA or TTT;
Z = A, T, or C;
XXXYYYZ ≄′ AAAAAAA or TTTTTTT.
Step 2: Search for a pseudoknot 3′ of the XXXYYYZ slippery site motif using the GenoBase program (Baher et al. 1992; Hagstrom et al. 1992). Constraints placed on the pseudoknot were as follows:
- 1.
- The pseudoknot must begin within 8 nucleotides of base Z;
- 2.
- Stem 1 must have a minimum length of 6 bp, containing no more than one mismatch, one insertion, and/or one deletion;
- 3.
- Gap 1 can be no greater than 3 nucleotides in length;
- 4.
- Stem 2 must have a minimum of 5 bp with only one insertion, deletion, or mismatch allowed;
- 5.
- Gap 2 can be no greater than 3 nucleotides in length;
- 6.
- Gap 3 is limited to 100 nucleotides in length.
Step 3: Align motifs found in steps 1 and 2 with an open reading frame (ORF) of at least 50 codons, such that the first base in the slippery site (the first X) is in the third base of a codon. Furthermore, searching in the 5′ direction of the motif there must be an in-frame ATG codon before a translational termination signal (TAA, TAG, or TGA). Sequences that satisfied all of these criteria were defined as “motif hits.”
Owing to the size of the individual yeast DNA sequences, that is, entire chromosomes, the yeast genomic database was divided into units of 104 bases and was subjected to the search protocol described above. Similarly, the human DNA database was divided into four different sections owing to its large size. The database output files in HTML format contain links to the complete GenBank entry for each locus, Medline links, protein links, nucleotide neighbors, structure links, and the complete locus sequence in FASTA format. The motif hits are described, and predicted peptide sequences for both −1 and 0 frames are provided. These were analyzed using BLAST searches (Altchul et al. 1990) to identify whether the motif hit occurred in a known gene. The outputs of the S. cerevisiae database search are available through the UMDNJ Department of Molecular Genetics and Microbiology World Wide Web page at http://www2.undnj.edu/mgenmweb/frameshifting.html. The other outputs are available upon request.
Strains, Media, Genetic Methods, and Plasmid Construction
E. coli strain DH5 was used for plasmid preparations, and transformations of E. coli and S. cerevisiae were performed as described previously (Dinman and Wickner 1992). YPAD and synthetic complete medium were as reported previously (Dinman and Wickner 1994). The S. cerevisiae strain JD88 (MAT a ura3-52 lys2-801 ade2-10 trp1Δ[L-AHNB] [M1]) was used for in vivo measurements of −1 ribosomal frameshifting efficiencies as described previously (Dinman and Wickner 1992). Yeast strain SJ2001 (MAT a leu2-3,112 ura3-52 his3Δ1 ade− trp1-289 ras1::HIS3 ras2::URA3[YEp13-TPK1]) was kindly provided by J. Broach (Princeton University, NJ). pRS306 (Sikorski and Hieter 1989) was digested with HindIII and BamHI, the overhanging ends were filled using Klenow fragment and dNTPs, and the resulting blunt ends were ligated together to make pJD171. pJD171 was then digested with PstI and EcoRV, creating a linear DNA fragment in which a significant portion of the URA3 gene was deleted. SJ2001 was transformed with the linearized pJD171, and Ura− colonies were selected for growth on medium containing 5-fluoro-orotic acid (5-FOA). The resulting strain, JD981, was used in the RAS1frameshifting studies.
The plasmids used in this study are shown in Figure 3A. pJD160.0 is derived from p314-JD86-ter (Cui et al. 1996), with the modification that it contains unique BamHI, SmaI, andKpnI restriction endonuclease recognition sites 3′ of the AUG start codon, and 5′ of the lacZ gene. This is the 0-frame control plasmid. pJD160.c-1 is identical to pJD160.0 except that lacZ is in the −1 frame with respect to the translational start site without any intervening frameshift signal. This is used to measure nonprogrammed −1 ribosomal frameshifting. p314-JD85-ter (Cui et al. 1996) is a programmed −1 ribosomal frameshift test plasmid that relies on the L-A frameshift signal to produce β-galactosidase activity. The RAS1 andCCR5 motif hits are depicted in Figure 3B. The frameshift signal from the yeast RAS1 gene was amplified from genomic DNA by PCR as described (Costa and Weiner 1995) using the synthetic oligonucleotide primers shown in Table 5. Because theRAS1 frameshift signal is predicted to direct ribosomes into premature termination signals, two additional nucleotides were added in the spacer regions between the slippery site and pseudoknot of these PCR products such that a −1 frameshift would redirect ribosomes into the original reading frame. The CCR5 frameshift signal was cloned by PCR from a CCR5 cDNA template [pBabe-CCR5, kindly provided by D. Littman, Skirball Institute (HHMI), New York, NY] using the synthetic oligonucleotide primers shown in Table 5. The respective PCR products were cloned into pJD160.0 to produce pJD160.RAS1 and pJD160.CCR5. In each of these constructs, a programmed −1 frameshift is required for the lacZ gene to be translated. Ribosomal frameshift efficiencies were calculated by dividing β-galactosidase activities from cells harboring test plasmids by β-galactosidase activities from cells harboring the 0-frame control plasmid and multiplying by 100% (Dinman et al. 1991).
Synthetic Oligonucleotides Used in this Study
A functional, full-length RAS1 gene was amplified by PCR from genomic yeast DNA using the oligonucleotides shown in Table 5, and the PCR products were cloned into pRS314 and pRS316 (TRP1 andURA3 CEN vectors, respectively) (Sikorski and Hieter 1989) to create pRAS1–TRP1 and pRAS1–URA3. JD981 cells were transformed with pRAS1–URA3 and selected for growth on medium lacking uracil (H−ura). The transformed cells were passaged in media lacking uracil only, and colonies were screened for loss of YEp13-TPK1 such that the Ras growth pathway was solely supported from Ras1p activity generated from pRAS1–URA3. Site-directed mutagenesis by PCR using the oligonucleotides shown in Table 5 was used to change the slippery site (wild type = GGGAAAT; amino acid sequence = Gly, Asn) in pRAS1–TRP1 to GGGCAAT or GGGTAAT to create pRAS1A → C.TRP1 and pRAS1A → T.TRP1 (see Fig. 3A). Cells harboring pRAS1–URA3 were transformed with these TRP1, and colonies were selected for growth on H−trp. Transformants were subsequently grown in the presence of 5-FOA to select for loss of pRAS1–URA3 (Rose et al. 1990). Cells harboring pRAS1–TRP1, pRAS1A → C.TRP1, and pRAS1A → T.TRP1 were subsequently tested for their abilities to use different carbon sources (2% dextrose, 3% ethanol, 3% glycerol, 2% potassium acetate) at 24°C, 30°C, and 37°C.
Acknowledgments
This work was supported by grants to J.D.D. from the National Institutes of Health (NIH) (R01 GM58859) and from The New Jersey Commission on Cancer Research (97-60-CCR). A.B.H. was supported in part by a training grant from the NIH (T32 AI07403-07). S.W.P. is supported by a grant from the NIH (R01 GM48631) and an Established Investigator Award from the American Heart Association.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL dinmanjd{at}umdnj.edu; FAX (732) 235-5223.
-
- Received January 4, 1999.
- Accepted February 24, 1999.
- Cold Spring Harbor Laboratory Press