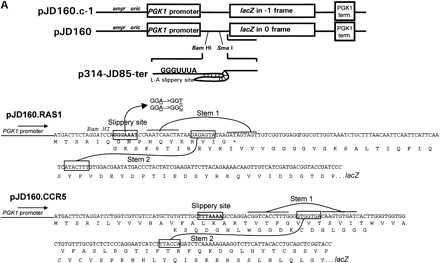
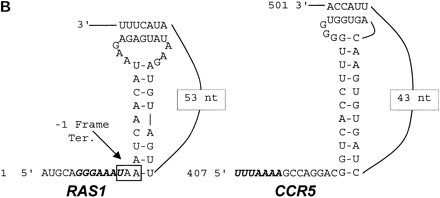
(A) Plasmids used to measure programmed −1 ribosomal frameshifting. pJD160.0 is the 0-frame control plasmid. pJD160.c-1 measures nonprogrammed −1 ribosomal frameshifting. p314-JD85-ter is used to measure L-A virus-promoted programmed −1 ribosomal frameshifting. The frameshift signal from the yeast RAS1 gene was cloned into pJD160 to produce pJD160.RAS1. Because theRAS1 frameshift signal is predicted to direct ribosomes into premature termination signals, two additional nucleotides were added in the spacer regions between the slippery site and pseudoknot of theRAS1 PCR product. The CCR5 frameshift signal was cloned from a CCR5 cDNA template into pJD160.0 to produce pJD160.CCR5. In each of these constructs, a programmed −1 frameshift is required for the lacZ gene to be translated. Ribosomal frameshift efficiencies were calculated by dividing β-galactosidase activities from cells harboring test plasmids by β-galactosidase activities from cells harboring the 0-frame control plasmid and multiplying by 100%. (B) Representations of the RAS1and CCR5 motif hits. Numbers refer to the 5′ and 3′ positions of the depicted nucleotides in the respective ORFs. Slippery sites are italicized in boldface. The RAS1 −1 frame termination codon is noted.











