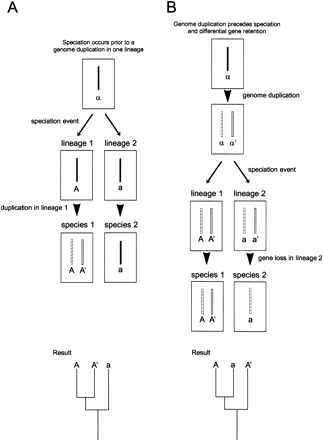
Two models explaining the origin of extra paralogs in one of two related species. (A) If a genome duplication occurs in lineage 1 after its divergence from lineage 2, then the resulting paralogous genomic elements (genes or chromosomes) A and A′ of an extant species from lineage 1 are more closely related to each other than they are to their ortholog a in an extant species from lineage 2. This relationship is evident in the resulting phylogenetic tree, diagramed at the bottom. (B) If a genome duplication produces paralogous genomic elements α and α′, then a subsequent speciation event will produce lineage 1 with paralogs A andA′ and lineage 2 with paralogs a anda′. A and a are orthologs, as they are both directly descended from α, and A′ anda′, descended from α′, are also orthologs. Lineage-specific loss of paralogs (e.g. a′ in lineage 2) results in species 2 having fewer paralogs than species 1. The derivation of A in species 1 and a in species 2 from their common ancestor α is evident in the resulting phylogenetic tree, diagramed at the bottom.











