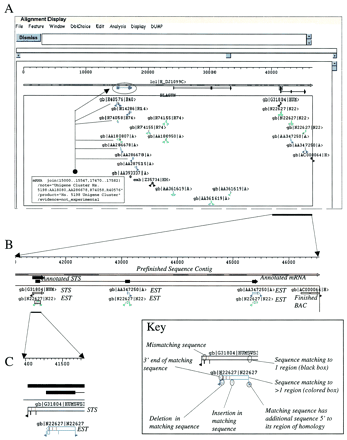
Three representative Musk views of the ASN.1 output generated for a single sequence contig by PowerBLAST from the lowest (A) to highest (C) resolution. Sequence matches to records in various databases are shown in a boxed region directly below the schematically depicted sequence contig. In addition, sequence annotations of the query sequence (e.g., Annotated mRNA) are shown as black lines and boxes immediately below the depicted query sequence. Regions in which the query sequence has been masked for repetitive elements are shown as crosshatched. (A) Global view of entire contig. Multiple sequence matches are shown within the boxed region labeled BLASTN. In addition, several user-added annotations (colored lines and boxes) are shown immediately below the contig sequence. In particular, a UniGene cluster has been annotated as an mRNA (blue). The ESTs corresponding to this hypothetical UniGene cluster are linked with dotted lines. (B) A higher resolution view of the last ∼6 kb of the sequence contig. Various sequence matches are shown, with additional text added to indicate the different types of matching sequences (e.g., EST, STS, finished BAC). Sequences matching to more than one region of the query sequence are colored to reflect the number of such matches. One EST (N22627) matches three regions (green), whereas another EST (AA347250) matches two regions (blue). Also indicated are the putative intron–exon boundaries for an mRNA containing several of the matching EST sequences, an STS (G31804), and the 5′ end of a finished BAC clone (AC000064). (C) A 200-bp region within the ∼6-kb segment shown in B. At this high level of detail, the degree of identity between the query sequence and matching sequences can be more clearly seen. Various such features are graphically indicated, including mismatches, insertions, and deletions (see the inset box for a key).











