A Prokaryotic Gene Cluster Involved in Synthesis of Lysine through the Amino Adipate Pathway: A Key to the Evolution of Amino Acid Biosynthesis
Abstract
In previous studies we determined the nucleotide sequence of the gene cluster containing lys20, hacA (lys4A),hacB (lys4B), orfE, orfF,rimK, argC, and argB of Thermus thermophilus, an extremely thermophilic bacterium. In this study, we characterized the role of each gene in the cluster by gene disruption and examined auxotrophy in the disruptants. All disruptants except for the orfE disruption showed a lysine auxotrophic phenotype. This was surprising because this cluster consists of genes coding for unrelated proteins based on their names, which had been tentatively designated by homology analysis. Although the newly found pathway contains α-aminoadipic acid as a lysine biosynthetic intermediate, this pathway is not the same as the eukaryotic one. When each of the gene products was phylogenetically analyzed, we found that genes evolutionarily-related to the lysine biosynthetic genes inT. thermophilus were all present in a hyperthermophilic and anaerobic archaeon, Pyrococcus horikoshii, and formed a gene cluster in a manner similar to that in T. thermophilus. Furthermore, this gene cluster was analogous in part to the present leucine and arginine biosyntheses pathways. This lysine biosynthesis cluster is assumed to be one of the origins of lysine biosynthesis and could therefore become a key to the evolution of amino acid biosynthesis.
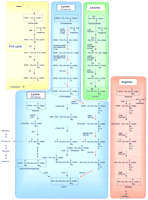
Lysine had been believed to be synthesized from aspartic acid through the diaminopimelic acid (DAP) pathway in all prokaryotic organisms. An extremely thermophilic Gram-negative bacterium, Thermus thermophilus, was shown to synthesize lysine not via DAP, but from 2-oxoglutaric acid and acetyl-CoA through the α-aminoadipic acid (AAA) pathway, (Fig. 1) which is known to be common in fungi (Kosuge and Hoshino 1998; Kobashi et al. 1999). Because higher eukaryotes other than animals synthesize lysine through DAP, the AAA pathway is a characteristic that distinguishes fungi from higher eukaryotes (Vogel 1964, 1965; Broquist 1971).
Comparison of biosynthetic pathways of lysine, leucine, and arginine, and a related part in the citric acid cycle. Red arrows indicate the putative AAA pathway of T. thermophilus and P. horikoshii.
In general, prokaryotes biosynthesize not only lysine but also methionine, threonine, and isoleucine from aspartic acid. T. thermophilus also possesses an aspartate kinase (Kobashi et al. 1999) that catalyzes the first reaction in the DAP pathway. However, this enzyme is only used for synthesis of methionine, threonine, and probably isoleucine. In T. thermophilus, lysine is synthesized through the AAA pathway (Kobashi et al. 1999; Kosuge and Hoshino 1998). This was the first discovery of lysine biosynthesis via AAA in bacteria. Here we report that the gene cluster for lysine biosynthesis through the AAA pathway is also found in a hyperthermophilic and anaerobic archaeon, Pyrococcus horikoshii, whose genome has been sequenced completely (Kawarabayasi et al. 1998), and discuss the evolution of amino acid biosynthesis.
RESULTS AND DISCUSSION
Lysine Biosynthetic Cluster in T. thermophilus
We characterized the role of each gene in the cluster by gene disruption (Kosuge and Hoshino 1997, 1998) and examined auxotrophy of the disruptants (Fig. 2). Because the genes in the cluster are closely arranged in the same orientation, the cluster seems to be an operon. Gene disruption by inserting a kanamycin-resistance gene could cause reduced expression of a distal gene, which might be required for lysine biosynthesis in an operon. At present, it is unclear whether or not the genes are transcribed in a single mRNA and have independent promoters. In any case, to weaken the possible polar effect, the kanamycin-resistance gene cassette and the promoter were inserted in the same orientation as the gene cluster, which may allow expression of the genes located downstream via a read-through transcript originated from the inserted promoter. All of the disruptants, except for those with orfE disruption, showed a lysine auxotrophic phenotype (Table 1). This shows that all of the genes except for orfE are involved in lysine biosynthesis. This was surprising because this cluster consists of genes coding for apparently unrelated proteins, as judged by their names, which were tentatively assigned by homology analysis. Considering the functions of the homologs of the gene products and auxotrophy of the disruptants, lys20 was shown to encode homocitrate synthase, and hacA and hacB to code for large and small subunits of homoaconitase, respectively (Kosuge and Hoshino 1998; Kobashi et al. 1999). Based on auxotrophy of the disruptants and the homology of the gene products, we assume that the ribosomal protein S6 modification enzyme (RimK),N-acetylglutamate kinase (ArgB), andN-acetylglutamate 5-semialdehyde dehydrogenase (ArgC) are involved in modification of AAA, phosphotransfer to the AAA derivative, and its reduction to yield an aldehyde compound, respectively (see below). This clearly indicates that the AAA pathway for lysine biosynthesis is used in prokaryotes as well as in eukaryotes, although some modification must be present. We also found the cluster of lysine biosynthetic genes in P. horikoshii but in no other organism in the international DNA/protein databases (EMBL, GenBank, and DDBJ).
Map of Km-resistance gene insertion point. Each of the pUCLU and pUCLD derivatives carrying the Km-resistance gene cassette at indicated positions was used for the disruption of the chromosomal copy ofT. thermophilus.
Effect of Gene Disruption on Growth of T. thermophilus
Phylogenetic Analysis of Components in the AAA Pathway ofT. thermophilus
The above results let us reconsider amino acid biosynthesis. We here phylogenetically analyze each protein encoded by the lys20,hacA, hacB, rimK, argC, andargB genes. The alignment data are available onhttp://iam.u-tokyo.ac.jp/misyst/lysclust.html.
Homocitrate Synthase of T. thermophilus
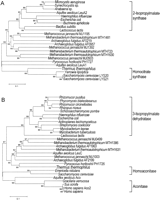
Homocitrate synthase catalyzes the reaction to form homocitric acid with 2-oxoglutaric acid and acetyl-CoA. This is the first reaction in the AAA pathway (Fig. 1). Homocitrate synthase of T. thermophilus (Lys20) is grouped with those of the yeastsSaccharomyces cerevisiae and Yarrowia lipolytica(Fig. 3A). In addition, this group is closely related to a protein, PH1727, of P. horikoshii and a protein, LeuA1, of a hyperthermophilic bacterium Aquifex aeolicus (Deckert et al. 1998). The lineage of homocitrate synthase is closely related to that of 2-isopropylmalate synthase (LeuA). LeuA is involved in the branched pathway toward leucine and catalyzes the reaction similar to that of homocitrate synthase, in which 2-isopropylmalic acid is used in place of 2-oxoglutaric acid as one of the substrates. The phylogenetic tree shows that homocitrate synthase and 2-isopropylmalate synthase share a common ancestor.
Comparison of TCA cycle, AAA lysine biosynthesis, and leucine biosynthesis related proteins. (A) Phylogenetic relationships among the amino acid sequence encoded in lys20 gene of T. thermophilus and high-scoring amino acid sequences from the result of homology search using PSI-BLAST (Altschul et al. 1997). A total of 332 amino acid sites were considered without gap regions in alignment. The bar indicates 10% difference of amino acid sequence. The DAD (DDBJ amino acid sequence database) accession nos. used in this phylogenetic analysis are D85684-1-1,D63999-62-1, Z46907-2-1, AE000772-5-1, U32779-6-1, AE000117-11-1,X71612-5-1, Z75208-75-1, U92974-14-1, U67561-1-1, AE000909-1-1,AE000922-3-1, U67500-4-1, AP000007-42-1, Z74230-1-1, Z74179-1-1,Z49114-1-1, AE000685-7-1, AE001091-11-1, U67579-2-1, AE000851-8-1, andAE001038-8-1. (B) Phylogenetic relationships among the amino acid sequence encoded in the hacA gene of T. thermophilus and the related sequences. A total of 279 amino acid sites were considered without gap regions in alignment. The DAD accession nos. used in this phylogenetic analysis are AP000007-41-1,AE000954-24-1, X99624-1-1, U46154-1-1, J05224-1-1, U87939-1-1,U80040-1-1, U17709-1-1, AE000756-5-1, AE000901-10-1, U67499-13-1,U67543-1-1, AE000967-2-1, AE000922-4-1, AE000714-13-1, U92974-16-1,Z99263-32-1, AL021287-2-1, AL031124-34-1, X84647-1-1, U32779-8-1,AE000117-9-1, Z99262-3-1, D67033-1-1, X53090-1-1, M33166-1-1, andD63833-1-1.
Homoaconitase of T. thermophilus
Homoaconitase (homoaconitate hydratase) catalyzes conversion ofcis-homoaconitic acid to either homocitric acid or homoisocitric acid, or vice versa. The large subunit of homoaconitase (HacA/Lys4A) of T. thermophilus is grouped with a protein, PH1726, of P. horikoshii (Fig. 3B). This group is connected to the lineage of 3-isopropylmalate dehydratase (LeuC) homologs fromA. aeolicus and three archaea, Archaeoglobus fulgidus(Klenk et al. 1997), Methanobacterium thermoautotrophicum(Smith et al. 1997) and Methanococcus jannaschii (Bult et al. 1996). Similar results were obtained with the small subunit, HacB/Lys4B, of homoaconitase (data not shown).
Each of these three archaea has two 3-isopropylmalate dehydratase homologs, both of which are also related to HacA of T. thermophilus and PH1726 of P. horikoshii. This may reflect the past existence of a common origin of homoaconitase and 3-isopropylmalate dehydratase in those microorganisms.
T. thermophilus homoaconitase consists of large and small subunits, HacA and HacB, like 3-isopropylmalate dehydratase, LeuC and LeuD, whereas fungal homoaconitase is a single, long polypeptide containing sequences for both large and small subunits. The environment of fungi has been quite different from the extremely thermophilic environment of Thermus and Pyrococcus. In addition, the phylogenetic tree (Fig. 3B) shows that T. thermophilushomoaconitase and its homolog of P. horikoshii share an ancestor with eukaryotic homoaconitases but that they possibly separated from the eukaryotic enzymes in the early stages of evolution. All the observations suggest that the homoaconitase gene was not transferred horizontally from prokaryotes to fungi.
The phylogenetic tree in Figure 3B contains aconitase in addition to homoaconitase and 3-isopropylmalate dehydratase. Aconitase (aconitate hydratase) reversibly catalyzes the conversion of cis-aconitic acid to either citric acid or isocitric acid in the citric acid cycle and therefore shares structural characteristics with homoaconitase and 3-isopropylmalate dehydratase. In this respect, recently Irvin and Bhattacharjee (1998) also reported that fungal homoaconitase shared a common ancestor with aconitase and 3-isopropylmalate dehydrogenase.
T. thermophilus RimK-Like Protein
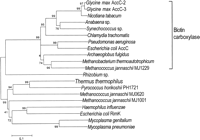
T. thermophilus contains a rimK-like gene within the gene cluster for lysine biosynthesis. The RimK protein ofEscherichia coli was isolated and characterized as a protein that adds glutamic acid residues to the carboxyl terminus of ribosomal protein S6 of the organism (Kang et al. 1989). The protein family including E. coli RimK is defined as a number of proteins with ATP-dependent carboxylate-amine ligase activity, where acylphosphate intermediates are involved in the catalytic mechanism (Galperin and Koonin 1997). The phylogenetic tree (Fig. 4) shows that there are two major groups: biotin carboxylase, and RimK. TheT. thermophilus RimK homolog is again most closely related toP. horikoshii PH1721. Other archaea, such as A. fulgidus, M. thermoautotrophicum, and M. jannaschii, have at least two proteins, each of which is classified to the biotin carboxylase or RimK groups. Interestingly, M. jannaschii has two homologs in the RimK subgroup: One is grouped with those of T. thermophilus and P. horikoshii, and the other is grouped with those including E. coli RimK. Considering that RimK-like protein of T. thermophilus is involved in lysine biosynthesis, the RimK homolog that is grouped with those of T. thermophilus and P. horikoshii might also have a similar catalytic activity required for lysine biosynthesis.
Phylogenetic relationships among the amino acid sequence encoded in therimK gene of T. thermophilus and the related sequences. A total of 164 amino acid sites were considered without gap regions in alignment. The bar indicates 10% difference of amino acid sequence. The DAD accession nos. used in this phylogenetic analysis areL14612-2-1, AE000404-10-1, AF007100-1-1, AF068249-1-1, L38260-1-1,L14862-1-1, U59234-2-1, AE000942-12-1, U67563-10-1, AE001090-1-1,AE001286-7-1, AP000007-36-1, U32828-12-1, X15859-2-1, U39679-12-1,AE000016-2-1, U67510-6-1, U67542-8-1, and AE000094-2-1.
T. thermophilus ArgC and ArgB
ArgC catalyzes the conversion of N-acetylglutamate 5-phosphate to N-acetylglutamate 5-semialdehyde. This is the third reaction in the arginine biosynthetic pathway. By homology search using PSI-BLAST (Altschul et al. 1997) for proteins homologous to that encoded by the argC gene in the lysine biosynthetic gene cluster of T. thermophilus, we found another gene having the name argC in the T. thermophilus genome (Baetens et al. 1998). This shows that T. thermophilus has at least twoargC homologous genes: One is used in arginine biosynthesis, and the other is used in lysine biosynthesis. Hereafter, we call the protein working in arginine biosynthesis as true-ArgC, and tentatively call the protein involved in lysine biosynthesis as pseudo-ArgC.
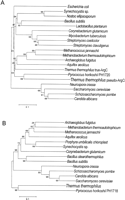
In the phylogenetic tree (Fig. 5A), T. thermophilus true-ArgC is grouped with an ArgC homolog of A. aeolicus and those of achaea, while pseudo-ArgC is most closely related to an ArgC homolog, PH1720, of P. horikoshii. P. horikoshii has no otherargC homolog in its complete genome sequence database. This suggests that ArgC of P. horikoshii is involved in both arginine and lysine biosyntheses. In any case, it is evident that T. thermophilus (and probablyP. horikoshii) lysine biosynthesis is related to the present arginine biosynthetic pathway.
Comparison of arginine biosynthesis and Thermus lysine biosynthesis related proteins. (A) Phylogenetic relationships among the amino acid sequence encoded in the argC gene ofT. thermophilus and the related sequences. A total of 267 amino acid sites were considered without gap regions in alignment. The bar indicates 10% difference of amino acid sequence. The DAD accession nos. used in this phylogenetic analysis are D64004-19-1, U48355-1-1,X52834-1-1, X99978-2-1, AL031541-2-1, M83659-1-1, Z85982-15-1,AF049897-1-1, M21446-1-1, AE000861-7-1, U67552-4-1, AE000961-28-1,AE000761-6-1, Y10525-2-1, AP000007-35-1, Z69727-9-1, X98880-1-1,U18813-19-1, and L27746-1-1. (B) Phylogenetic relationships among the amino acid sequence encoded in the argB gene ofT. thermophilus and the related sequences. A total of 157 amino acid sites were considered without gap regions in alignment. The DAD accession nos. used in this phylogenetic analysis areAE001016-12-1, AE000806-5-1, U67464-11-1, AE000771-3-1, M94625-2-1,D90905-74-1, X86157-3-1, L06036-3-1, Z26919-3-1, X98880-1-1,U18813-19-1, L27746-1-1, Z69727-9-1, and AP000007-33-1.
The phylogenetic tree for ArgB of T. thermophilus is shown in Figure 5B. The tree shows that ArgB of T. thermophilus is also closely related to its homolog PH1718 in P. horikoshii. TheT. thermophilus and P. horikoshii lineage is far from both prokaryotic and eukaryotic ArgBs which catalyze conversion ofN-acetylglutamate to N-acetylglutamate 5-phosphate in the arginine biosynthetic pathway. P. horikoshii has a single ArgB homolog as is the case with ArgC.
In the arginine biosynthetic pathway, N-acetylglutamate is converted to N-acetylglutamate 5-semialdehyde viaN-acetylglutamate 5-phosphate, which is a series of the reactions catalyzed by ArgB and ArgC. On the other hand, in the lysine biosynthesis pathway of eukaryotes, 2-aminoadipic acid is converted to 2-aminoadipic acid 6-semialdehyde via an adenosylated derivative (Fig.1) by a multifunctional enzyme, aminoadipate reductase (Morris and Jinks-Robertson 1991; Miller and Bhattacharjee 1996). No significant similarity in amino acid sequence is observed between aminoadipate reductase and ArgB/C. Because the lysine biosynthetic pathway inT. thermophilus is suggested to proceed in a mechanism similar to that in arginine biosynthesis, we assume that a 2-aminoadipic acid derivative possibly modified by a RimK homolog is converted to a 6-semialdehyde derivative by pseudo-ArgC and ArgB in the lysine biosynthetic gene cluster. Thus, the lysine biosynthetic pathway inT. thermophilus may not be completely the same as that in the eukaryotes.
Horizontal Gene Transfer Between Thermusand Pyrococcus?
As described above, all the enzymes encoded in the lysine biosynthetic gene cluster in T. thermophilus are closely related to the proteins in P. horikoshii. Surprisingly, the corresponding genes in P. horikoshii are grouped in a manner similar to T. thermophilus (Fig. 6). Here, we mention the possibility of horizontal gene-cluster transfer betweenT. thermophilus and P. horikoshii, each of which live in a hyper-thermogenic environment, because only T. thermophilus and P. horikoshii have similar lysine biosynthetic gene clusters among the 19 organisms examined. All the phylogenetic trees show that the gene cluster for lysine biosynthesis in T. thermophilus and P. horikoshii are closely related to each other.
Comparison of T. thermophilus gene cluster for lysine biosynthesis and the corresponding region in P. horikoshii. The percentages show amino acid sequence identity between the homologous genes.
However, the trees also indicate that the branching point is deep, probably close to the early divergence of bacteria and archaea. Considering that the earliest organisms had lived under hot and anaerobic conditions, we assume that this gene cluster preserves one of the origins of lysine biosynthesis and that the lysine biosynthetic gene cluster has been independently conserved in both organisms. Nelson et al. (1999) reported that 24% of the predicted coding sequences in the Thermotoga maritima genome were most similar to proteins in archaeal species, and almost half were similar to P. horikoshii. However, T. maritima does not have the gene cluster for AAA lysine biosynthesis but has the lysA gene for DAP lysine biosynthesis.
Amino Acid Biosynthesis of P. horikoshii
Although P. horikoshii does not have a gene manipulation system, its amino acid biosynthesis can be deduced by analysis of phylogenetic trees and by use of genetic evidence from T. thermophilus, whose lysine gene organization is similar. Diaminopimelic acid decarboxylase (LysA) catalyzesmeso-2,6-diaminopimelic acid to lysine. This is the last reaction in the biosynthesis from aspartic acid to lysine (Fig. 1). The homology search using PSI-BLAST reveals that P. horikoshii has no LysA homolog. This is consistent with the idea that P. horikoshii synthesizes lysine not through DAP, but via the AAA pathway like T. thermophilus. When the gene clusters are compared between T. thermophilus and P. horikoshii, both organisms possess a single additional gene, orfE andPH1722, in the cluster (Fig. 6). We do not know the function of OrfE. However, the amino acid sequence of OrfE is homologous to that of OrfF, which is required for lysine biosynthesis in T. thermophilus. Interestingly, both OrfE and OrfF are composed of repetitive short segments of 20–30 amino acids, all of which contained a C-P/E-x-C-G motif. We speculate that gene duplication oforfF may have occurred. PH1722 has high amino acid sequence similarity to 3-isopropylmalate dehydrogenase (LeuB) and isocitrate dehydrogenase (data not shown). By analogy of the reactions mediated by these enzymes, we assume that PH1722 possesses homoisocitrate dehydrogenase activity. P. horikoshii possesses no other homologs of PH1722, PH1724, PH1726, and PH1727 in its genome. This suggests that these four proteins might be involved in leucine biosynthesis as LeuB, LeuD, LeuC, and LeuA, respectively, as well as lysine biosynthesis, as P. horikoshii was reported to show only Trp auxotrophy (Gonzalez et al. 1998). It should be noted that these enzymes are structurally similar to enzymes consisting of a part of the citric acid cycle (Fig. 1). Considering that the earliest organisms are believed to have lived under hot, anaerobic conditions, it is reasonable to postulate that amino acid biosynthesis developed earlier than the citric acid cycle, which functions under aerobic conditions. It is possible that the aconitase of the tricarboxylic acid (TCA) cycle evolved from homoaconitase or 3-isopropylmalate dehydratase.
In addition to the four genes PH1722, PH1724,PH1726, and PH1727, P. horikoshii has singleargC and argB homologs in its genome. Furthermore, the archaeon has argD (PH1716) and argE(PH1715) homologs, which may have the activities of aminotransferase and deacylase just downstream of the putativeargB gene PH1718. By analogy to arginine biosynthesis, we assume that the semialdehyde-type AAA derivative formed by PH1720 and PH1718 could be converted to lysine by the ArgD and ArgE homologs in P. horikoshii. The organism contains only a single copy of argC, argB, argD, andargE homologs in its genome. We therefore speculate that the products of these genes may possess the activity required for arginine biosynthesis.
Although P. horikoshii and T. thermophilus have very similar clusters of lysine biosynthetic genes, it is unlikely that these organisms have identical synthetic pathways, because T. thermophilus has at least two argC homologs while P. horikoshii has a single argC homolog. This suggests that lysine and arginine biosyntheses are separated in T. thermophilus. Consistent with this, the argB disruptant ofT. thermophilus requires only lysine for its growth, but not arginine (Table 1). In this study, it is suggested that P. horikoshii might have developed unique amino acid biosynthetic systems in which several amino acids are synthesized by a limited number of enzymes with broad substrate specificity. The presence of lysine biosynthesis through AAA is suggested for an extremely thermophilic anaerobic archaeon, Thermoproteus neutrophilus, based on acetate assimilation patterns (Schäfer et al. 1989). It is therefore interesting to elucidate the lysine biosynthetic pathway of T. neutrophilus.
Evolution of Amino Acid Biosynthesis
P. horikoshii grows optimally under anaerobic conditions at a temperature of nearly 100°C. Considering that the earliest organisms had lived under hot and anaerobic conditions,Pyrococcus may preserve prototypic proteins. The enzyme recruitment model for the evolution of metabolic pathways suggests the enzymes which are somewhat loose in their substrate specificity could initially function in multiple pathways and evolve late into two or more specific enzymes by gene duplication (Jensen 1976). In fact, several enzymes from Pyrococcus are reported to have broad substrate specificity (Kengen at al. 1993; Bauer et al. 1996; Fischer et al. 1996; Mai and Adams 1996; Durbecq et al. 1997; Glasemacher et al. 1997). In this study we propose that P. horikoshii, likeT. thermophilus, synthesizes lysine through the AAA pathway. We further suggest that unlike T. thermophilus, at least four of these lysine genes are also involved in leucine synthesis and four other genes in that cluster are involved in arginine synthesis. This system suggested for P. horikoshii could be one of the earliest amino acid biosynthetic systems, in which enzymes recognized at least two different substrates.
Interestingly, the other archaea, A. fulgidus, M. thermoautotrophicum, and M. jannaschii, each have two or three homologs of lys20, hacA, and hacB. The phylogenetic trees (Fig. 2A,B) show that each gene was duplicated or triplicated at an early stage of evolution, probably before the species diverged. At present, we do not know the homologs' physiological substrates. However, it may be possible to speculate that some of them are still in a stage of evolution seeking for enhancing substrate specificity. Pyrococcus has lived on the earth for a long time under hot and anaerobic conditions, which may allow the organism to have some prototypic enzymes. In this study we propose a hypothesis that the present biosynthetic pathways for lysine, leucine, and arginine could have developed by ancestor enzymes with broad substrate specificity. We believe that further studies on the amino acid biosynthetic systems in P. horikoshii will help elucidate the evolution of amino acid biosynthesis.
METHODS
T. thermophilus was transformed as described (Koyama et al. 1986). Disruption of each gene was performed according to the method of Kosuge and Hoshino (1997, 1998). Each disruption was confirmed by Southern hybridization. The detailed data are available on request.
We performed a homology search using PSI-BLAST (Altschul et al. 1997) with the given parameter values on the DNA data bank of Japan (DDBJ). This program compares a given query amino acid sequence against all other proteins in the databases to identify related sequences. Here the query amino acid sequences were AAA lysine biosynthesis-related proteins from T. thermophilus. Each of the multiple alignments was created using the CLUSTAL W program (Thompson et al. 1994) on DDBJ among a query sequence and high-scoring sequences of the PSI-BLAST result. The phylogenetic tree from p-distance estimation by the neighbor-joining method (Saitou and Nei 1987) with 1000 bootstrap analyses (Felsenstein 1985) was constructed based on the multiple alignment using MEGA version 1.01 (Kumar et al. 1993).
Acknowledgments
We thank Dr. Tetsuo Hamamoto for providing helpful comments for completing the draft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE
We renamed lys20, hacA (lys4A),hacB (lys4B), orfE, orfF,rimK, argC, and argB in the cluster ofT. thermophilus, as lysS, lysT,lysU, lysV, lysW, lysX,lysY, and lysZ, respectively, because the cluster is actually involved in lysine biosynthesis.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL umanis{at}mail.ecc.u-tokyo.ac.jp; FAX 81-3-5841-8030.
-
- Received July 13, 1999.
- Accepted October 14, 1999.
- Cold Spring Harbor Laboratory Press