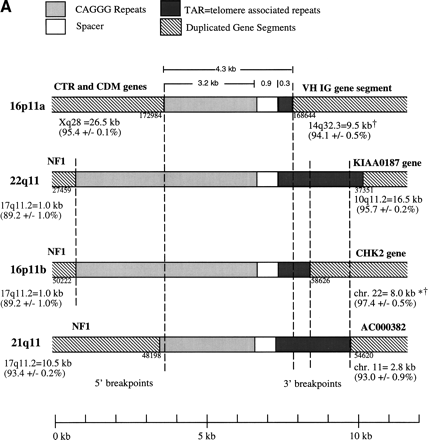
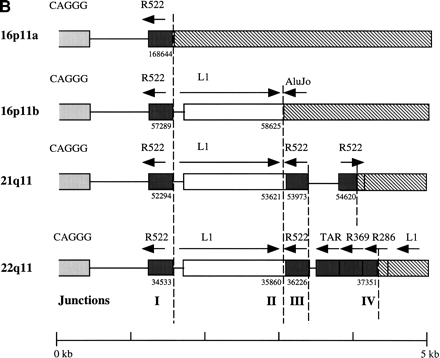
Duplicated genomic segments flank the CAGGG repeat structure. (A) Four human CAGGG repeat elements [21q11, 22q11, and two from 16p11 (a and b)] were identified based on BLAST sequence similarity searches of htgs and nr divisions of GenBank (accession nos.AC004527, D87003, AC002041, and AC002402, respectively). Multiple pairwise sequence alignments (CLUSTA W) were used to determine the extent of the repeat structure. The diagram shows the structure of the repeat element, composed of (1) an interspersed CAGGG repeat block (mean length = 4490 bp; range, 2937–5915 bp), (2) a short spacer region (mean length = 881 bp; range, 851–943), and (3) a variety of subtelomeric-like repeat sequences (HSREP522, HSREP271, TAR, HSREP282) (mean length = 1899 bp; range, 793–3018 bp). Broken lines represent breakpoints within the aligned repeat units. Analysis of sequences flanking the repeat structure identified paralogous gene structures located near or at the boundaries of each repeat. The duplicated genic segments are shown as hatched bars. Numbers below these boundaries refer to the breakpoint positions within the GenBank accession. The percent sequence similarity, the location of the ancestral sequence, and the estimated size of the duplication are summarized below each bar. (*) Calculation of sequence similarity was based on sim4 comparisons of cDNA and genomic segment. (†)Minimal size of duplicated region was based on the genomic distance between first and last paralogous exons. All interchromosomal duplication boundaries were confirmed by PCR amplification and sequencing of products from a monochromosomal somatic cell hybrid panel. (B) The location of the 3′ boundary of the repeat element with respect to various telomere-like repeat elements is shown in greater detail. Note the preponderance of breakpoints near or within the HSREP522 (R522) element.











