
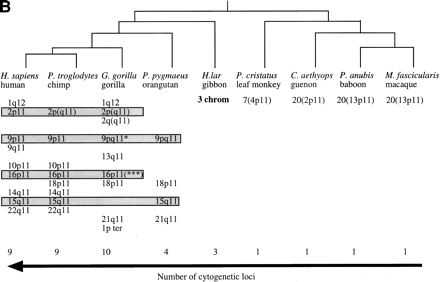
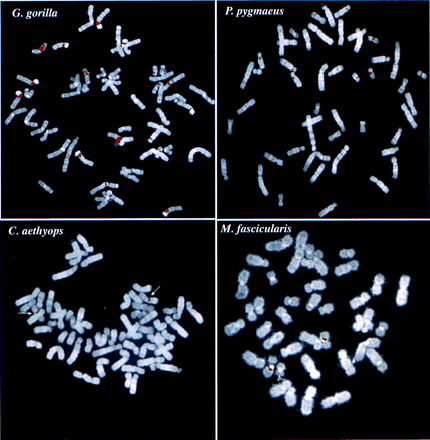

Evolutionary conservation and distribution of the CAGGG repeat. (a) A 1.9-kb subclone (p196.3.12) was constructed within the CAGGG interspersed repeat sequences that flank the CTR-CDM (creatine-transporter/DXS1357E) duplicated locus on 16p11.2 (Fig. 1). FISH analysis performed on a human metaphase chromosomal spread identified nine pericentromeric cytogenetic localizations (1q12, 2p11, 9q11, 9q12, 10p11, 14q11, 15q11, 16q11, and 22q11). (b) A summary of comparative FISH analysis using the same probe on metaphase chromosomal spreads from nine different primate species. The locations of these signals with respect to human phylogenetic group assignment are presented in the context of a generally accepted phylogeny of these species. Note that hominoid species show a general increase in the number of chromosomal sites that hybridize with this probe. The human phylogenetic assignments among the cercopithecoids were confirmed by using whole or partial human chromosome painting probes (species-specific band assignments are shown in parentheses). (*) The correspondence to human 9p11 or 9q11 cannot be unambiguously resolved because of recent pericentric inversion. (***) Three distinct hybridization signals could be distinguished on GGO chromosome 17q11 (HSA 16p11 equivalent). (c) Comparative FISH analysis of CAGGG repeat probe, 196.3.12. Representative hybridizations of chromosomal metaphases are shown for two hominoids (gorilla and orangutan, G. gorilla and P. pygmaeus) and two Old World monkey species (guenon and macaque,C. aethyops and M. fascicularis). (d) A sequence alignment of a portion (∼350 bp) of the CAGGG repeat structure from two baboon loci (denoted BAB followed by BAC coordinate; GenBank accession nos. AF144085 and AF144086) and two human (HUM; GenBank accession nos. AC004527 and AC002041) loci. Regions of sequence identity are shaded.











