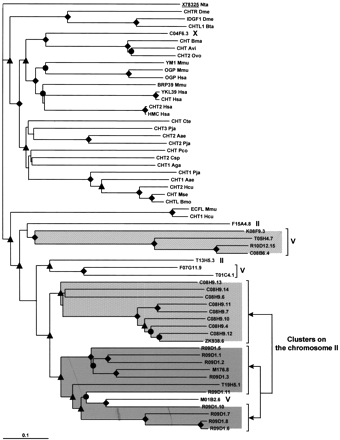
Phylogenetic analysis of chitinase-like proteins of C. elegans. Sequences analyzed in this tree are identical to those listed in Table 3 except for eight sequences that show an incomplete chitinase domain and have not been used in the phylogenetic analysis. Chromosomal localizations of C. elegans chitinase genes are indicated at right; the labeled shaded box indicate the two clusters localized on chromosome II (see Fig. 1). Symbols for bootstrap values are as in Fig. 3. The sequence X78325 (underlined) ofNicotiana tabacum is used as outgoup. Species abbreviations used for the phylogenetic tree are as follows: (Aea) Aedes aegypti (yellow fever mosquito); (Aga) Anopheles gambiae(african malaria mosquito); (Avi) Acanthocheilonema viteae(animal filarial nematode); (Bmo) Bombyx mori (silkmoth); (Bta) Bos taurus; (Bma) Brugia malayi (agent of Brugian lymphatic filariasis); (Bpa) Brugia pahangi (filarial nematode of domestic cats); (Csp) Chelonus sp. (braconid wasp); (Cte) Chironomus tentan (nonbiting midges); (Dme)Drosophila melanogaster (fruit fly); (Hcu) Hyphantria cunea (fall webworm); (Hsa) Homo sapiens (human); (Mse)Manduca sexta (tobacco hornworm); (Mmu) Mus musculus(mouse); (Nta) Nicotiana tabacum (common tobacco); (Ovo)Onchocerca volvulus (agent of river blindness); (Pco)Phaedon cochleariae (mustard beetle); (Pja) Penaeus japonicus (Kuruma prawn); (Wba) Wuchereria bancrofti(agent of lymphatic filariae).











