Genomics and Hearing Impairment
Abstract
Hearing impairment is clinically and genetically heterogeneous. There are >400 disorders in which hearing impairment is a characteristic of the syndrome, and family studies demonstrate that there are at least 30 autosomal loci for nonsyndromic hearing impairment. The genes that have been identified encode diaphanous (HDIA1), α-tectorin (TECTA), the transcription factorPOU4F3, connexin 26 (GJB2), and two unconventional myosins (MYO7A and MYO15), and four novel proteins (PDS,COCH, DFNA5, DFNB9). The same clinical phenotype in hearing-impaired individuals, even those within the same family, can result from mutations in different genes. Conversely, mutations in the same gene can result in a variety of clinical phenotypes with different modes of inheritance. For example, mutations in the gene encoding MYO7A cause Usher syndrome type IB, autosomal-recessive nonsyndromic hearing impairment (DFNB2), and autosomal-dominant nonsyndromic hearing impairment (DFNA11). Additionally, the mouse ortholog of theMYO7A gene is the shaker-1 gene. Mouse models such asshaker-1 have facilitated the identification of genes that cause hearing impairment in humans. The availability of high-resolution maps of the human and mouse genomes and new technologies for gene identification are advancing molecular understanding of hearing impairment and the complex mechanisms of the auditory system.
Advances in genomics have been instrumental in clarifying the genetic heterogeneity of nonsyndromic hearing impairment. As of November 1998, the Hereditary Hearing Loss Homepage (URL: http://dnalab-www.uia.ac.be/dnalab/hhh) (Van Camp and Smith 1998) lists the autosomal locations of 19 loci for dominant (DFNA*) and 19 for recessive (DFNB*) nonsyndromic sensorineural hearing impairment (Fig. 1). The genes at 13 of these autosomal loci have been identified; DFNA1,DFNA8 (also DFNA12), DFNA15, DFNB1(also DFNA3), DFNB2 (also DFNA11), andDFNB3 encode the highly conserved diaphanous (HDIA1) (Lynch et al. 1997), α-tectorin (TECTA) (Verhoeven et al. 1998), the transcription factor POU4F3 (Vahava et al. 1998), the gap junction protein connexin 26 (GJB2) (Kelsell et al. 1997; Denoyelle et al. 1998), the unconventional myosin VIIA (MYO7A) (Liu et al. 1997a,b; Weil et al. 1997), and a second unconventional myosin (MYO15) (Wang et al. 1998), respectively. The genes identified at the remaining four loci (DFNA5, DFNA9,DFNB4, and DFNB9) encode novel proteins (Li et al. 1998; Robertson et al. 1998; Van Camp et al. 1998; Van Camp and Smith 1998). Mutations in two of these genes, MYO7A andDFNB4, were shown to cause Usher syndrome type IB (Weil et al. 1995) and Pendred (PDS) syndrome (Everett et al. 1997), respectively, before being also associated with nonsyndromic hearing impairment. The characteristic findings in Usher syndrome type I patients are profound sensorineural hearing impairment from birth, progressive retinal degeneration, and vestibular dysfunction, whereas patients with Pendred syndrome have thyroid goiter in addition to congenital sensorineural hearing impairment.
Chromosomal locations of genes for hearing impairment. Numbers atleft of the chromosome give the cytogenetic band assignments for each of the loci. Those genes that have been identified are denoted by asterisks and the encoded protein is shown below the chromosome. Mouse genes that are known to be, or likely to be, orthologous are shown to the left and human genes to the right. DFNB15 is in parentheses because it is equally likely to be on chromosomes 3 and 19, and USH1F is in parentheses because its cytogenetic location on chromosome 10 is unknown. [Constructed from the Hereditary Hearing Loss Home Page (Van Camp and Smith 1998)].
History
Documentation of awareness that inheritance is important in hearing impairment can be traced back to the sixteenth century. According toGoldstein (1933), the earliest known author to have recognized that some forms of deafness may be hereditary was Johannes Schenck (1531–1598) who noted a family in which several children were born deaf. Stephens (1985) includes a pedigree drawing of a sixteenth century family of the Spanish aristrocracy in which members in three generations were documented as deaf. In 1621 the papal physician Paolus Zacchias (1584–1659) recommended that the deaf abstain from marriage because of evidence that their children will also be deaf (Cranefield and Federn 1970), indicating his conviction that heredity is important in deafness. Reardon (1990) ascribes to Sir William Wilde (1815–1876) the awareness that deafness shows different patterns of inheritance, that consanguinity is a relevant factor, and that there is an excess of males among the congenitally deaf. These findings were confirmed byHartmann (1881) who carried out extensive studies in schools for the deaf in Germany.
More recently, Konigsmark (1969), Konigsmark and Gorlin (1976), andFraser (1976) provided comprehensive reviews of hereditary hearing impairment, and emphasized the pronounced heterogeneity. The phenotypic and genetic heterogeneity is underscored by Gorlin et al. (1995) who list 427 forms of syndromic and nonsyndromic hereditary hearing impairment. Several studies attempted to estimate the number of loci for deafness in various populations (Stevenson and Cheeseman 1956;Chung et al. 1959; Sank 1963; Chung and Brown 1970; Costeff and Dar 1980; Brownstein et al. 1991) with the results ranging from less than ten to several thousand. As far as pattern of inheritance is concerned, there is general agreement that ∼77% is autosomal recessive, 22% is autosomal dominant, and the remainder are X-linked and mitochondrial (Gorlin et al. 1995). Fraser (1976) pointed out that inheritance pattern is useful but not sufficient for categorizing type of hearing impairment and he predicted that “within the foreseeable future it will be possible to define each type of genetically determined deafness by other criteria, such as can be employed at present, for example, for defining galactosemia and phenylketonuria, on biochemical grounds as two specific forms of mental subnormality.” Twenty years later this prediction is becoming a reality.
Syndromic vs. Nonsyndromic Hearing Impairment
Clinically, hearing impairment may be associated with other disorders (syndromic) or it may be the only symptom (nonsyndromic). However, both syndromic and nonsyndromic phenotypes can result from mutations in the same gene (e.g., MYO7A, PDS). Also, more extensive examination of nonsyndromic patients may reveal subclinical defects in other organs or tissues. Syndromic hearing impairment tends to be less genetically heterogeneous than nonsyndromic, but more than one locus has been identified for several syndromes. In fact, the number of loci that have been mapped so far for Usher syndrome type I (USH1*) is six, and some families do not show linkage to any of those loci. Figure 1 shows that USH1C(Smith et al. 1992) maps to the same location as DFNB18 (Jain et al. 1998) on the short arm of chromosome 11, and USH1D(Wayne et al. 1996) and DFNB12 (Chaib et al. 1996) are both in the same region of the long arm of chromosome 10. The identification of these genes may provide additional examples of different mutations in the same gene causing both syndromic and nonsyndromic hearing impairment.
Other syndromes for which more than one gene has been identified include Waardenburg, Stickler, Alport, and Jervell and Lange-Nielsen. Many different mutations in a paired-box gene, PAX3, and a transcription factor, MITF, result in the characteristic clinical findings of type I and type II Waardenburg syndrome (WS) (Tassabehji et al. 1995). Mutations in the procollagen 2 gene (COL2A1) on chromosome 12q13 were shown to cause Stickler syndrome (STL) (Williams et al. 1996). However, ∼50% of families do not show linkage to COL2A1, and mutations in COL11A1on chromosome 1p21 and COL11A2 on chromosome 6p21 have been found in some of these families (Richards et al. 1996; Vikkula et al. 1995). Types II and XI collagen have been shown to copolymerize within collagen fibrils (Mendler et al. 1989) and are necessary for skeletal development. Collagen is also defective in Alport syndrome, which is associated with mutations in genes encoding type IV collagen. A defective gene on the X chromosome (COL4A5) causes most cases of Alport syndrome (Barker et al. 1990). However, mutations in the genes that encode two other subunits of type IV collagen are known to cause autosomal-recessive Alport syndrome. These genes, COL4A3and COL4A4, are located on the long arm of chromosome 2 (Mochizuki et al. 1994). Each of these type IV collagens is found in the basilar membrane, parts of the spiral ligament, and stria vascularis, but how defects in these collagens result in sensorineural hearing loss has not yet been elucidated. In Jervell and Lange-Nielsen syndrome (JLNS), congenital profound sensorineural hearing loss is associated with an electrocardiogram that has a prolonged Q–T interval. The two genes in which mutations have been identified,KVLQT1 on chromosome 11p15.5 (Neyroud et al. 1997) andKCNE1 on chromosome 21q22 (Tyson et al. 1997), both encode proteins that form the delayed rectifier potassium channel, which is necessary for endolymph homeostasis within the ear. Mutations in these genes also cause Ward-Romano syndrome, an autosomal-dominant disorder in which affected individuals have a prolonged Q–T interval but they are not hearing impaired (Wang et al. 1996; Splawski et al. 1997).
Genetic Epidemiology
Approximately one in a thousand children is born with profound hearing impairment and at least 70% have no associated anomalies (Gorlin et al. 1995). Analysis of data collected by Gallaudet University in the 1969–1970 Annual Survey of Hearing Impaired Children and Youth estimated that 50.7% of cases are attributable to genetic causes (Nance et al. 1977), whereas analysis of the 1988–1989 data estimated a genetic etiology in 62.8% of cases (Marazita et al. 1993). A major reason for the increase in the percentage caused by genetic factors is the reduction in environmental causes (such as rubella). However, an underlying genetic predisposition is likely to be relevant in the majority of cases in which environmental factors are known to have played a role.
Adult-onset hearing loss is also a significant health problem, with 14% of individuals between the ages of 45 and 64 and 30% of those older than 65 years having hearing problems (Hotchkiss 1989). Sill et al. (1994) collected extensive population data in an attempt to delineate the causes of later-onset hearing loss. The data strongly suggested a genetic etiology in a large percentage of the participants, but a precise estimate could not be determined.
Age-dependent penetrances need to be incorporated into the genetic analysis of later-onset hearing loss. Additionally, the likely possibility of phenocopies must be taken into account in the analysis of family data, regardless of age of onset. An example of this within-family heterogeneity was found in the DFNA15 kindred (Vahava et al. 1998). One member with hearing loss showed recombination with markers on chromosome 5. The pattern of hearing loss in this individual was different from that in other affected members, and mutational analysis of the POU4F3 gene demonstrated that this individual does not have the mutation associated with hearing loss in other members of the family.
Phenotypes and Populations
Severe-to-profound hearing impairment with onset before 12 months of age is a characteristic of the affected members of most of the families for which genes causing recessive hearing impairment have been localized. These families are from endogamous populations and/or the affected individuals are the offspring of consanguineous matings. The countries of origin include Pakistan, India, Tunisia, Bali, Lebanon, Palestine, Israel, and Syria. An exception to preverbal onset is a large consanguineous kindred from Pakistan in which affected individuals had normal hearing until ∼10 yr of age but the loss became profound within 4–5 yr (Veske et al. 1996). This locus (DFNB8) was mapped to the long arm of chromosome 21. In general, loci for hearing impairment in unrelated families have been mapped to different chromosomal regions. However, mutations in theGJB2 gene on chromosome 13 have been found in affected individuals from many ethnic backgrounds, and connexin 26 defects may explain >50% of autosomal-recessive deafness (Zelante et al. 1997). Additionally, the same GJB2 mutation (30delG) is found in a large percentage of these individuals (Denoyelle et al. 1997). However, a different mutation (167delT) probably accounts for the majority of cases of nonsyndromic recessive deafness in the Ashkenazi Jewish population (Morell et al. 1998).
Most loci for autosomal-dominant hearing impairment have been mapped in single large pedigrees. However, independent studies of families in Belgium and Austria localized DFNA12 and DFNA8 to 11q22–q24, and mutations in the TECTA gene have been found in affected members of both families (Verhoeven et al. 1998). Also, several unrelated families show linkage to DFNA2 on the short arm of chromosome 1 (Van Camp et al. 1997; Talebizadeh et al. 1998) and three different families are linked to DFNA10 on the long arm of chromosome 6 (O’Neill et al. 1996), suggesting that defects in the same gene are responsible for the hearing loss. The onset of the hearing loss in families with a dominant mode of inheritance is generally after the development of speech, and the frequencies affected tend to be consistent among members of the same family. An exception is the hearing loss in members of the DFNA3 family in which the onset is before 4 yr of age and is severe-to-profound across all frequencies (Chaib et al. 1994). DFNA3, which maps to the same region of chromosome 13 as DFNB1, is also caused by mutations in theGJB2 gene (Denoyelle et al. 1998).
Sometimes ancestry may allow prediction of the location of the disease gene. For example, all families of Acadian ancestry with Usher syndrome type I map to a narrow region on the short arm of chromosome 11. The majority of the affected individuals are homozygous for the same haplotype over a 6-cM interval, suggesting a single mutation (Keats et al. 1994). This haplotype has not been found on Acadian chromosomes that do not have the disease allele. Thus, for profoundly hearing impaired infants of Acadian ancestry, marker typing can differentiate between nonsyndromic hearing impairment and Usher syndrome type IC.
Relevance of Advances in Genomics
The development of high-resolution genetic and physical maps, together with the construction of genomic and cDNA libraries, and the availability of sequence databases for many species, have provided the tools for finding genes for hearing impairment. A human fetal cochlear cDNA library (Robertson et al. 1994) has been valuable in the cloning of many of the hearing-impairment genes so far identified, in particular, DFNA9 (Robertson et al. 1998). By studying sets of related hearing impaired individuals who are likely to have the same defective gene, DNA pooling strategies (Sheffield et al. 1994) and a genome screen have been used effectively for genetic mapping. In most cases, this approach has led to a single location for the defective gene. However, DFNB15 is equally likely to be in two locations, one on chromosome 3 and the other on chromosome 19, based on the analysis of three affected and three unaffected offspring of first cousins (Chen et al. 1997). The maximum lod score for both locations is 2.78 and additional data are required to determine if both, one, or neither loci are responsible for the deafness in this family. This study demonstrates the necessity of performing a complete genome search, especially in small, consanguineous families.
The usual strategy for localizing a hearing impairment locus in a family is to begin by genotyping markers in the vicinity of all of the mapped loci. Using this approach, several of the loci for dominant and recessive forms of hearing impairment have been mapped to the same regions (Fig. 1), suggesting that the same gene may be responsible. Two examples have already been confirmed: the hearing impairment in both the DFNB2 and DFNA11 kindreds is caused by MYO7A mutations, and mutations in GJB2 are responsible for the hearing impairment in DFNA3 and DFNB1 kindreds.
Although X-linked and mitochondrial forms of nonsyndromic hearing impairment are much less common than autosomal forms, advances in genome research have facilitated the identification of defective X-linked and mitochondrial genes in these forms of hearing impairment. As might be expected, the first nonsyndromic hearing impairment locus mapped was in families showing X-linked inheritance (Brunner et al. 1988; Wallis et al. 1988). This mixed (conductive and sensorineural) hearing impairment is associated with perilymphatic gushers if stapedectomy is done. The defective gene (DFN3) encodes the transcription factor POU3F4 (de Kok et al. 1995). Three other nonsyndromic hearing-impairment genes have been localized on the X-chromosome (Fig. 1) but they have not yet been identified.
Hearing impairment is often one of the components of the phenotype in mitochondrial myopathies that are caused by specific mitochondrial DNA (mtDNA) mutations (Fischel-Ghodsian 1998). In some families hearing impairment may be the only anomaly. An mtDNA mutation (A1555G) in the 12S rRNA gene was found to be the cause of hearing impairment in a family showing a maternal pattern of inheritance. This same mutation is associated with aminoglycoside-induced hearing loss (Prezant et al. 1993) and estimates of the frequency of this mutation suggest that screening hearing-impaired individuals for this mutation may be beneficial in some populations (Estivill et al. 1998). Another form of hearing loss that may be associated with mtDNA mutations is presbycusis. Fischel-Ghodsian et al. (1997) found a significant increase in mtDNA mutations in the temporal bones of patients with presbycusis and evidence is mounting that the accumulation of mtDNA mutations may be relevant to many aspects of the aging process (Nagley et al. 1993).
Mouse Models
Comparative genomics has made many valuable contributions to disease gene identification (Meisler 1996), and hearing impairment is no exception (Brown and Steel 1994). A major advantage of using the mouse for finding disease genes is the relative ease of obtaining large numbers of informative progeny. A relevant example is the humanUSH1B locus and the mouse deafness mutant shaker-1(sh1), which were hypothesized to be caused by mutations in orthologous genes because they had been mapped to a conserved linkage group on human chromosome 11q13 and mouse chromosome 7. This hypothesis was proven to be correct when Gibson et al. (1995) showed that thesh1 gene encoded myosin VIIa, and Weil et al. (1995) quickly found mutations in the human MYO7A gene in Usher type IB patients. Similarly, the mouse mutant shaker-2 (sh2) and the deafness in kindred DFNB3 are both caused by mutations in another unconventional myosin (MYO15) and complementary work in mice and humans led to this finding (Probst et al. 1998; Wang et al. 1998).
Both of the genes for Waardenburg syndrome were identified because of previous studies in mice. Mapping of the genes for Waardenburg syndrome types I and II to chromosomes 2 (Farrer et al. 1992) and 3 (Hughes et al. 1994) led to their identification as the human homologs of the mouse genes that cause the splotch mutant (Epstein et al. 1991; Goulding et al. 1991), and the microphthalmia mutant (Tassabehji et al. 1994), respectively.
The POU4F3 gene became a candidate for the dominant hearing impairment in the DFNA15 family when it was mapped to a region of the long arm of chromosome 5 that shares homology with the region on mouse chromosome 18 containing the Pou4f3 (also known asBrn3c and Brn-3.1) gene. Targeted deletion of both alleles of Pou4f3 had been shown to cause deafness in mice (Erkman et al. 1996; Xiang et al. 1997). Although the critical region on chromosome 5 was too large (25 cM) to begin a physical mapping approach to finding the gene, the hunch that POU4F3 may be the defective gene proved to be true (Vahava et al. 1998).
No human loci have yet been mapped to the orthologous regions of mouse chromosomes 9 and 6 containing the Snell’s waltzer(sv) and deafwaddler (dfw) loci, respectively. The sv gene encodes an unconventional myosin heavy chain, myosin VI (myo6) (Avraham et al. 1995) and thedfw gene encodes a plasma membrane Ca2+-ATPase type 2 pump (Atp2b2) (Street et al. 1998). Both of these mouse mutants are deaf and show the typical head-tossing behavior of mice with inner ear defects, and it seems likely that hearing impaired patients with defects in these genes will be found eventually.
Another likely ortholog of a human gene for hearing impairment is thedeafness (dn) gene on mouse chromosome 19 (Keats et al. 1995). This region shares homology with human chromosome 9q13–q21 to which both DFNB7 and DFNB11 have been mapped (Jain et al. 1995; Scott et al. 1996).
Auditory Pathway and Protein Function
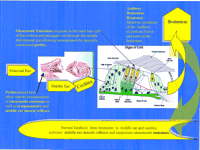
Studying endogamous populations, consanguineous matings, and large pedigrees minimizes heterogeneity but does not necessarily eliminate it. In some cases auditory testing may detect phenotype differences among members of the same kindred. Such findings may be the result of etiological differences and provide critical information for genetic studies. Figure 2 shows a flow diagram of the auditory pathway linking the external ear to the cochlea to the brainstem. The auditory tests that can be done to determine the location of the hearing deficit in patients are provided in Table 1. For example, the pure tone audiogram plots the lowest intensity at which a patient responds against the frequency of a pure tone that is placed into the ear for up to several seconds. Both air and bone are used for conduction of the sound, and if both audiograms are abnormal the hearing loss is sensorineural. However, if the bone conduction audiogram is normal or substantially better than the air conduction audiogram, then the loss is conductive and caused by a problem in the middle ear which is for the most part bypassed with bone-conducted sound. Abnormal tympanometry (a mechanical test of middle ear impedance) as well as elevation or elimination of middle-ear muscle reflex responses provide confirmation of a conductive loss.
Expected Test Results for a Selection of Putative Locations of Common Hearing Deficits
A more precise categorization of sensorineural hearing loss can be made by testing otoacoustic emissions, which are sounds coming from the motion of the outer hair cells. They allow cochlear function to be assessed independently of neural function, and recent studies (Morell et al. 1998) suggest that they may be helpful in discriminating between those who carry certain mutations that cause recessive nonsyndromic sensorineural hearing impairment and those who do not. In some patients, otoacoustic emissions may be normal but the auditory brainstem response (ABR) which monitors synchrony of the auditory (cochlear) nerve may be abnormal or absent, providing differentiation between a sensory and a neural loss (Starr et al. 1996; Berlin et al. 1998). Thus, extensive auditory testing provides critical information for determining the location of the hearing deficit and consequently detecting phenocopies in family data. Sophisticated analyses of ABR and otoacoustic emission data may eventually allow prediction of the gene defect that is causing the hearing impairment.
Most forms of hearing impairment involve abnormal development of the outer and inner hair cells (sensory cells) in the cochlea (Fig. 2). The human ear has ∼3600 inner hair cells and 3.5 times as many outer hair cells. The outer hair cells are structurally well-organized and are motile. The inner hair cells do not move, but both types of cells have an upper surface of w-shaped stereocilia that consist of an actin core and a myosin outer cover, and are joined to one another by tip links. In response to sound vibrations, the stereocilia bend and pull on the tip links, and this movement opens ionic channels that allow transduction of mechanical to electrical energy within the hair cells. Intricate to this process is the communication between the stereocilia of the hair cells and the tectorial membrane, which is an extracellular matrix in close proximity to the hair cells. Note that the inner hair cells are not touching the tectorial membrane, whereas the outer hair cells are connected to the tectorial membrane. This mechanical arrangement presumably allows the motile outer hair cells to control, filter, and/or amplify motion to the stereocilia of the inner hair cells (Brownell et al. 1985). Additionally, ion channels and gap junctions are critical for the movement of potassium ions from the hair cells to the endolymph such that the correct electrical signal is sent to the auditory nerve.
Scanning electron microscopy shows that stereocilia are malformed when the MYO7A, MYO15, and MYO6 genes are defective, suggesting that these myosins are essential for maintaining the structure of the stereocilia (Steel and Brown 1998). Diaphanous may also be necessary for establishing the rigid structure of the actin core of the stereocilia (Lynch et al. 1997). The protein, TECTA, on the other hand, is a component of the tectorial membrane. Abnormal formation of either the hair cells or the tectorial membrane would be likely to disrupt the auditory pathway (Fig. 2). Similarly, if the channels or the pumps that allow passage of ions in and out of the hair cells are defective, then endolymph homeostasis is disrupted and the sound energy cannot reach the brain. Mutations in KVLQT1,KCNE1, GJB2, and Atp2b2 would all be expected to interfere with ion transport, and thus lead to hearing impairment.
DISCUSSION
Phenotypic and genetic heterogeneity are challenging realities that must be confronted to advance understanding of hearing impairment. Many different loci cause hearing impairment and the etiology may be different even among family members. As well as locus heterogeneity, allelic heterogeneity is a common finding. This allelic heterogeneity is sometimes associated with clinical heterogeneity, with both syndromic and nonsyndromic hearing impairment being caused by mutations within the same gene. Also, different mutations within the same gene cause dominant and recessive forms of hearing impairment. In contrast, distinct mutations within the same gene and different genes may give the same clinical phenotype, as is found for recessive nonsyndromic hearing impairment. Thus, the problem of useful categorization based on the clinical phenotype and mode of inheritance has been clarified substantially by the localization, and in some cases, identification of the gene, and genetic counseling for hearing impairment can now include the option of molecular genetic diagnostic testing. Such testing, particularly for GJB2 mutations, may provide the etiological answer for the parents of a profoundly hearing impaired child with no family history of hearing problems.
Identification of genes for hearing impairment provides an important step towards understanding the molecular mechanism of hearing. Environmental factors such as noise, infections, and drugs that contribute to hearing loss have been documented extensively, and delineating the effects on hearing of genetic and environmental factors and the interactions between them is progressing. The mtDNA mutation that predisposes to aminoglycoside-induced hearing loss is an obvious example of the interaction of genes and environment in etiology of hearing loss. In some cases, a single-gene defect may be the major contributor to the hearing impairment, in others it may be an environmental insult. But they are just the initiating factors; they set in motion a cascade of cellular events that inhibit the ability of the cell to function correctly. Advances in genomics have facilitated rapid progress in identifying genes for hearing impairment, and consequently increased insight into the normal development and function of cells in the auditory system.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL biombjk{at}lsumc.edu; FAX (504) 568-8500.
- Cold Spring Harbor Laboratory Press