Radiation Hybrid Mapping of the Two Highly Homologous Human–Variant pMCHL Genes by PCR–SSCP
Abstract
When gene loci are very similar in sequence, as in gene families or multiple pseudogenes, it is difficult to determine the specific location of the individual genes. We show here that applying PCR–SSCP to a radiation hybrid panel allowed mapping and specific sequencing of two genes with only a few sequence differences. Human–variant forms of the promelanin-concentrating hormone (pMCH) gene are found in two locations in the genome, previously localized by FISH to 5p14 and 5q12–q13. Without prior knowledge of sequence variation between the loci, we observed a difference in migration pattern in PCR–SSCP, indicating the presence of at least one point of sequence divergence. PCR–SSCP of 93 samples from a human–hamster radiation hybrid panel revealed the location of the genes to be between markers WI-4804 and AFM225YC5 on chromosome 5p, and between markers WI-3133 and WI-4225 on chromosome 5q. Sequencing of the two 680-bp PCR products from the hybrid panel demonstrated 3 bases of sequence difference between the 5p and 5q locations.
[The data described in this paper for thepMCHL2 gene have been submitted to the GenBank data library under accession no. AF064698.]
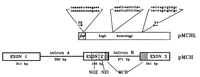
The rat pMCH gene codes for peptide precursor molecules that are processed into melanin-concentrating hormone (MCH) and other peptides (Parks and Vale 1992; Toumaniantz et al. 1996). Whereas the rat genome contains one copy of this gene, a human pMCH probe revealed three human loci in FISH mapping (Pedetour et al. 1994). Locus-specific probes showed that 12q23.1 is the location of the human homolog of the rat pMCHgene. The other two locations, on 5p14 and 5q12–q13, were visualized with a probe from a human–variant pMCH-like clone (pMCHL) that did not hybridize to the 12q location. Both chromosome 5 loci yielded equivalent hybridization signals, making it impossible to infer which of these sites corresponded to the sequence of the clone (Pedetour et al. 1994). The sequence of the human–variant clone was homologous to pMCH only within exons 2 and 3 and the intervening intron but diverged from the 12q pMCH sequence upstream of exon 2 (Breton et al. 1993). No homolog for exon 1 ofpMCH has been reported for these loci. However, the sequence for the peptide MCH is contained within exons 2 and 3 and could theoretically still be expressed. Therefore, these 5p and 5q sites were termed human–variant pMCHL.
The fine-map locations of the human–variant pMCHL loci are of interest because a region on 5p has been reported to be linked to bipolar disorder (Kelsoe et al. 1996) and because of the possible association of schizophrenia with trisomy of 5q11–q13 (Bassett et al. 1988). Furthermore, physiological data indicate that the peptide MCH could be considered a candidate for the neurochemical basis of psychiatric illness. In an animal model of an auditory processing deficit found in schizophrenia and in the manic phase of bipolar disorder, MCH administration induces the auditory evoked potentials seen in humans with the disease state (Miller et al. 1993).
The goal of our work was to determine whether sequence differences exist between these two sites of human–variant pMCHL using PCR–SSCP (Orita et al. 1989), a method sensitive enough to distinguish single points of sequence difference. If differences were found, they could be used to fine map and sequence each locus. For fine mapping, we used a 93-cell panel of human × hamster radiation hybrids (RHs) (Gyapay et al. 1996). To our knowledge, this is the first time PCR–SSCP has been used to fine map and sequence two highly homologous gene regions from a RH panel.
RESULTS
Initial PCR–SSCP screening for human–variant pMCHLusing chromosomal DNA from 14 individuals indicated the presence of two pairs of pMCHL bands. The likelihood that this represented the heterozygous state of a genetic polymorphism present in all individuals was low. Rather, it appeared more likely that the separate pairs of bands corresponded to the 5p and 5q loci of the pMCH gene and that the migration differences seen on the SSCP gel could be used to map the two genes separately.
Application of PCR–SSCP to the GB4 RH Panel
The results confirmed that the SSCP polymorphism discriminated two loci and not two alleles of the same locus. Each of 93 human/hamster hybrid samples were amplified using the pMCH variant-specific primer pair, run on an SSCP gel, and scored for the presence or absence of each band pair (Fig. 1). A hamster gene was also amplified with these primers (top band in Fig. 1), which provided an internal control for random PCR failure. Five possible scorings were obtained: both human bands amplified, only band 1 of human, only band 2 of human, no human bands but positive hamster band, and no human or hamster bands. In nine samples, the concomitant absence of a hamster product and absence of a human product prompted a rerun of the PCR for those samples with more DNA (25 ng rather than 12.5 ng). In six of the samples that were rerun, one of the human bands was subsequently identified on an SSCP gel.
Image of SSCP gel showing samples positive for a top pair of human–variant pMCH bands, for a bottom pair of bands, and for both pairs of bands (lanes 3 and 7, 8and 9, and 2 and G, respectively). Lane G contains the results of PCR amplification of the human–variant pMCHfrom the full complement of genomic DNA from a single individual. The background hamster product is noted at the top of the gel. On an agarose gel, the hamster band runs at ∼2300 bp and the human bands are 680 bp. The asterisk marks lane 1, an example of an RH sample that was negative for both hamster and human product.
The results for the lower band pair of Figure 1 placed one human–variant locus for pMCHL on 5p between markers WI-4804 and AFM225YC5, 13.35 cR from WI-4804, lod > 3, and identical to marker AFM225YC5, except for two values that are likely false positives because their existence at this site would require a double recombination event between marker WI-4804 and AFM225YC5. The number of matches and mismatches were as follows: The score for the lower band pair of pMCHL in 80 hybrids was identical with both flanking markers, in 11 hybrids the score was identical to AFM225YC5 but not WI-4804, and in 2 hybrids the score was discordant with both flanking markers. The other human–variant locus for pMCHL (the top band pair in Fig. 1) lies on 5q between markers WI-3133 and WI-4225, 19.24 cR from WI-3133, and closest to WI-4225. The number of matches and mismatches were as follows: The score for the top band pair ofpMCHL in 80 hybrids was identical with both flanking markers, in 11 hybrids the score was identical to WI 4225 but not WI 3133, in 1 hybrid the score was identical to WI-3133 but not WI-4225, and in 1 the score was discordant with both flanking markers. All data have been submitted to the Radiation Hybrid Data Base at EBI (http://www.ebi.ac.uk/RHdb).
Sequencing of the Human–Variant 5p and 5q pMCHL Genes
Samples of the GB4 panel positive for one or the other locus (but not both) were reamplified using the high-fidelity PCR enzyme Pwo. Manual and automated sequencing of the products was performed following gel purification. The DNA sequence of the lower band (Fig. 1), corresponding to the 5p locus, was identical to the sequence published for the human–variant clone (pMCH10 in Breton et al. 1993). The DNA sequence of the top band, corresponding to the 5q locus, exhibits three sequence differences (capitalized in Fig.2) across the 680-bp region amplified by the primer pair used in this study: one at position 737 (see Breton et al. 1993) in the begining of (putative) exon 2, where a C is found instead of an A; one at position 957 in intron B, where a G is found instead of an A; and the third at position 1212 in exon 3, where an A is found instead of a C. Additional sequence differences within 20 bp of each primer cannot be ruled out, as clear sequence of these regions was not obtained.
Organization of the pMCH gene, illustrating the region of sequence overlap for the pMCH gene on 12q and the human–variant pMCHL genes on 5p/5q, and showing the sites of sequence difference between the 5p and 5q human–variant loci. The 5p sequence is presented above the 5q sequence, with the points of difference in uppercase letters. Also shown are the primers used forpMCHL-specific PCR amplification (P2 and P7). The region with slanted lines indicates the coding region for the peptide—neuropeptide glycine–glutamic acid (NGE). The region with horizontal lines indicates the coding region for the peptide—neuropeptide glutamic acid–isoleucine (NEI). The regions with open circles indicate the coding region for MCH. It is not known whether the exon/intron structure is conserved in the region of homology between pMCHand pMCHL genes.
DISCUSSION
PCR–SSCP of pMCHL1 and pMCHL2 from the GB4 RH panel allowed fine mapping of these highly homologous genes. These mapping data are consistent with but more precise than previous FISH mapping data (Pedetour et al. 1994). In addition, selective sequencing from the RH DNA allowed us to determine the sequence differences between the two genes. The previously published pMCHL sequence (Breton et al. 1993) matches the sequence reported here forpMCHL1 of the 5p locus and is different from the 5q locus,pMCHL2, by only 3 bp of sequence across a region spanning ∼650 bp.
Prior work indicated that at least one of the pMCHL loci is expressed in the human CNS (Breton et al. 1993). However, the available data made it impossible to determine whether it is pMCHL1, on 5p, or pMCHL2, on 5q, that is expressed. This would be important, for example, when evaluating these loci as candidate genes for human psychiatric disorders. Knowledge of the sequence difference between the two loci greatly facilitated our subsequent transcription studies in which we show that the 5p locus is expressed (Miller et al. 1998). PCR–SSCP and selective sequencing of RHs has potentially broad applications for similar cases where several highly homologous genes or pseudogenes are present in the genome. This strategy provides a rapid means of determining whether sequence differences are present, what the sequence differences are, and which sequence is correlated with a precise chromosomal location. Other potential applications include degenerate PCR of gene families or the separation of mouse and hamster bands in rodent RHs.
METHODS
Primer Selection
Selection of the PCR primers was carried out with the Primer 1 program on the WI-MIT website, designed to span as much of the known sequence as possible. The sequence of the 5′ (P2) and the 3′ primer (P7) were, respectively, agaggaaggggtactggcct and atgggcttctcctcccattg.
PCR Conditions
The 10× PCR buffer composition was as follows: 100 mm Tris (pH 9.2), 50 mm NH4Cl, 27.5 mm MgCl2, 375 mm KCl, 0.1% porcine skin gel, and 50% glycerol. One picomole of the forward primer and 1 pmole of the reverse primer were used in each reaction. The dNTP concentration in the reaction was 400 μmoles for each dNTP, including dCTP. The quantity of Taq polymerase added to each reaction was 0.5 units. An amount equal to 0.5 μCuries of [32P] dCTP (stock 10 μCi/μl; Amersham) was added to each PCR reaction. All reactions were carried out in a total volume of 10 μl and overlaid with one drop of molecular biology grade mineral oil (Sigma). The PCR program was as follows: 94°C for 4 min, followed by 10 cycles of (93°C for 30 sec, 62°C for 2 min, 72°C for 2 min) and then 18 cycles of (93°C for 30 sec, 55°C for 2 min, 72°C for 2 min) with a final 5 min extension at 72°C.
SSCP Conditions
Conditions for SSCP were modified from those of Orita et al. (1989), using a nondenaturing gel of 4.3% acrylamide in 0.5× TBE, run at 30 W for 1.5 hr in a 4°C cold room.
RH Panel
The human–hamster RH panel (93 samples) was supplied by Research Genetics Genebridge 4 (Gyapay et al. 1996). The amount of each sample used in our experiments was 12.5 ng of DNA per PCR reaction.
Data Analysis
Each band pair was analyzed separately, by entering its presence or absence in each sample of the RH panel into the matrix provided by the Whitehead Institute/MIT Center for Genome Research Mapping Service (http://www-genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl). The algorithm at that website then generated the fine-map locations.
Sequencing
To generate products for sequencing, PCR amplification was carried out under conditions to minimize polymerase error, using a high-fidelity thermostable DNA polymerase system (0.5 units:Pwo + Taq in the Expand system supplied by Boehringer Mannheim), and a lower concentration of dNTPs (200 μm) than that used for the screening PCR (400 μm). Two bands, representing the two strands, are seen for each allele on an SSCP gel. For each band, two separate panel samples containing only that allele were PCR amplified, gel purified in low melting T agarose (GIBCO), extracted, and sequenced separately. Isolated PCR bands were sequenced using both manual (U.S. Biochemical) and automated methods (Applied Biosystems).
Acknowledgments
C.M. was supported by a training grant from the National Institute of Deafness and Other Communication Disorders (T32 DC00011).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL margit{at}umich.edu; FAX (734) 647-4130.
-
- Received February 23, 1998.
- Accepted May 22, 1998.
- Cold Spring Harbor Laboratory Press