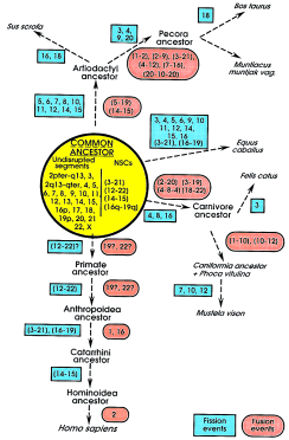
Schematic representation of the proposed karyotypic fission/fusion events in the evolution from the common ancestor to the seven species investigated and the primates (see text for references), which most likely resulted in the formation of the respective karyotypes. The events represent only those that could be identified (or can be proposed) based on the comparative painting data and therefore do not represent other possible rearrangements. Because in each case, correspondence with relation to individual human chromosomes was studied, all of the numbers presented relate to human chromosomes. In the common ancestor (yellow circle), two categories of chromosomes were included. Undisrupted segments represent those human chromosomes or chromosomal arms, the homologs of which were found to be entirely conserved in at least one of the seven species. HSA1p and 1q may also be added to this group; NSCs represented human chromosomes that were found to be contiguously syntenic in at least five species. Beginning from the common ancestor, the probable fission (blue rectangles) and fusion events (red ellipses) leading to the karyotypes of the respective species can be followed along the arrows. Commencing from the proposed ancestral configuration, the comparative painting data indicate very few major rearrangements (2–3 fusion and 3–5 fission events) leading to the formation of the cat, harbor seal, American mink, and human karyotypes; on the other hand, several obvious rearrangements (13–19 fission and 2–5 fusion events) leading to the pig, cattle, Indian muntjac, and horse karyotypes can be observed.











