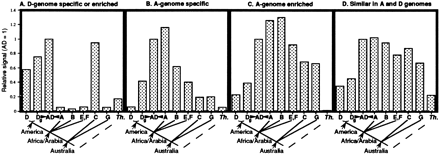
Densitometry analysis of dispersed repetitive DNA hybridization inGossypium. Labeled insert DNA from each of the 83 repetitive DNA clones was hybridized to slot-blotted total genomic DNA of 20 different cotton species, representing each of the 7 recognized genome types and the AD tetraploids (as listed in Fig. 1 legend, except that Dg designates G. gossypioides, and Th. designates the outgroup Thespesia lampas). For each of the 83 repetitive DNA probes applied to slot-blotted DNA of each of the 20 cotton species, signal was quantified by densitometry, standardized relative to the average signal of the two AD-genome species (defined as 1.0), and adjusted for genome size ratios as indicated in Methods to make results directly comparable. Average signal intensities across the 83 probes are plotted for each Gossypium genome type. Gossypiumgenome types are grouped according to our present understanding ofGossypium phylogeny based on chloroplast DNA restriction site variation (Wendel and Albert 1992). Geographic distributions for taxa are also indicated.











