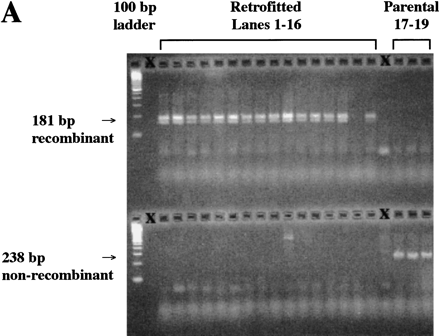
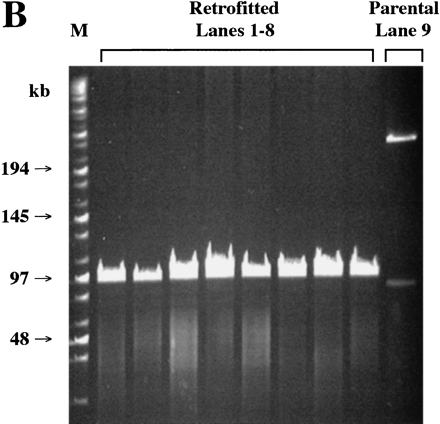
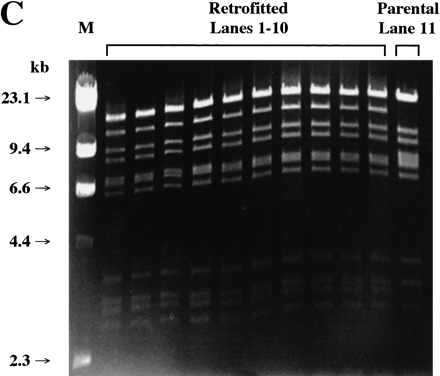
Analysis of randomly selected colonies obtained after recombination of BAC clone 261M17 and RETRObac. (A) Whole cell PCR of retrofitted clones (lanes 1–16) and parental BAC clones (lanes 17–19). (Top) Of 16 clones, 15 are positive for an expected 181-bp recombined product using primers Bac-F and Lac-R, whereas all parental clones are negative. (Bottom) Only the parental clones are positive for a 238-bp nonrecombined product using primers Bac-F and Bac-R. (X) Skipped lanes. (B) PFGE after AscI restriction digest of retrofitted clones (lanes 1–8) or the parental BAC clone (lane9). All eight retrofitted clones show a single band of ∼100 kb, whereas the parental clone is undigested by AscI. (C) Fingerprint analysis after HindIII restriction digest of retrofitted clones (lanes 1–10) or the parental BAC clone (lane 11). All 10 retrofitted clones show identical bands, whereas the parental clone has three differences.











